Back
Abstract Session
Session: Abstracts: Pediatric Rheumatology – Clinical II: JIA (2209–2214)
2213: Multi-omic Analysis of Macrophage Activation Syndrome Associated with sJIA Reveals a Potential Role of Type I Interferons in the Expansion of Cycling T Cells
Monday, November 14, 2022
4:00 PM – 4:10 PM Eastern Time
Location: Room 119
- PL
Pui Lee, MD, PhD
Boston Children's Hospital
Newton, MA, United States
Presenting Author(s)
Kailey Brodeur1, Liang Chen1, zhengping huang2, Yan Du1, Holly Wobma3, Maria Taylor4, Joyce Chang3, Megan Day-Lewis3, Fatma Dedeoglu3, Olha Halyabar3, Mindy Lo3, Jane W. Newburger5, Mary Beth F. Son3, Robert Sundel3, Peter Nigrovic3, lauren henderson3 and Pui Lee3, 1Boston Children's Hospital, Boston, MA, 2Guangdong Second Provincial Hospital, Guangzhou, China, 3Division of Immunology, Boston Children's Hospital, Boston, MA, 4Division of Immunology, Boston Children's Hospital, Brighton, MA, 5Department of Cardiology, Boston Children's Hospital, Harvard Medical School, Boston, MA
Background/Purpose: Macrophage activation syndrome (MAS) is a complication of systemic juvenile idiopathic arthritis (sJIA) characterized by cytokine storm and overt immune cell activation. We aim to further understand the immunologic landscape of sJIA and MAS.
Methods: We profiled peripheral blood mononuclear cells (PBMCs) from patients with active sJIA with or without MAS at the time of sampling and age-matched controls using bulk RNA sequencing (RNA-seq), single-cell RNA-seq, and mass cytometry. All patients with sJIA met ILAR diagnostic criteria and MAS was defined according to the 2016 Classification criteria for MAS complicating sJIA. Findings from RNA-seq and mass cytometry were validated using additional samples from patients with sJIA as well as patients with other inflammatory diseases by flow cytometry and in vitro T cell stimulation studies.
Results: Gene set enrichment analysis of bulk RNA-seq of PBMCs from patients with MAS (n = 9) revealed strong expression of genes associated with type I interferon signaling and T cell proliferation in addition to the expected type II IFN (IFN-g) signature based on gene set enrichment analysis. These features were rarely seen in age-matched healthy controls (n = 18) or patients with sJIA without MAS (n = 15). Single-cell RNA-seq confirmed the presence of type I and type II IFN signatures on all major PBMC subsets in patients with MAS (n = 8) while the T cell proliferation signature was localized to a population of Ki67+ cycling T cells with enhanced expression of IFN-g and genes involved in glycolysis, mTOR signaling and cytotoxicity (Figure 1).
Mass cytometry revealed that the cycling T cell population comprised of predominantly CD8+CD38+HLA-DR+ T cells and fewer CD4+CD38+HLA-DR+ T cells (Figure 2). We found that an expansion of CD38+HLA-DR+ T cells beyond 9% of CD8+ T cells or 4% of CD4+ T cells distinguished MAS from active sJIA, while a milder increase was generally seen in patients with sJIA without MAS compared to healthy controls (Figure 3). The degree of CD8+CD38+HLA-DR+ T cell expansion was also significantly greater in MAS compared to multi-system inflammatory syndrome in children (MIS-C) associated with COVID-19 (n = 17), acute viral infections (n = 15), Kawasaki disease (n = 7) and systemic lupus erythematosus (n = 9). In addition, stimulation of isolated T cells from healthy donors with IFN-I, but not IFN-II, augmented the effects of IL-12, IL-15 and IL-18 to generate CD8+CD38+HLA-DR+ T cells in vitro, while Janus kinase inhibition by ruxolitinib mitigated the impact of IFN-I treatment.
Conclusion: Our multi-omic analysis further details the expansion of the recently described CD8+CD38+HLADR+ T cells in MAS associated with sJIA. Our findings demonstrate a synergistic role of IFN-I in the generation of these cycling T cells and provide a mechanistic link between viral infections and MAS that may be targetable by JAK inhibition.
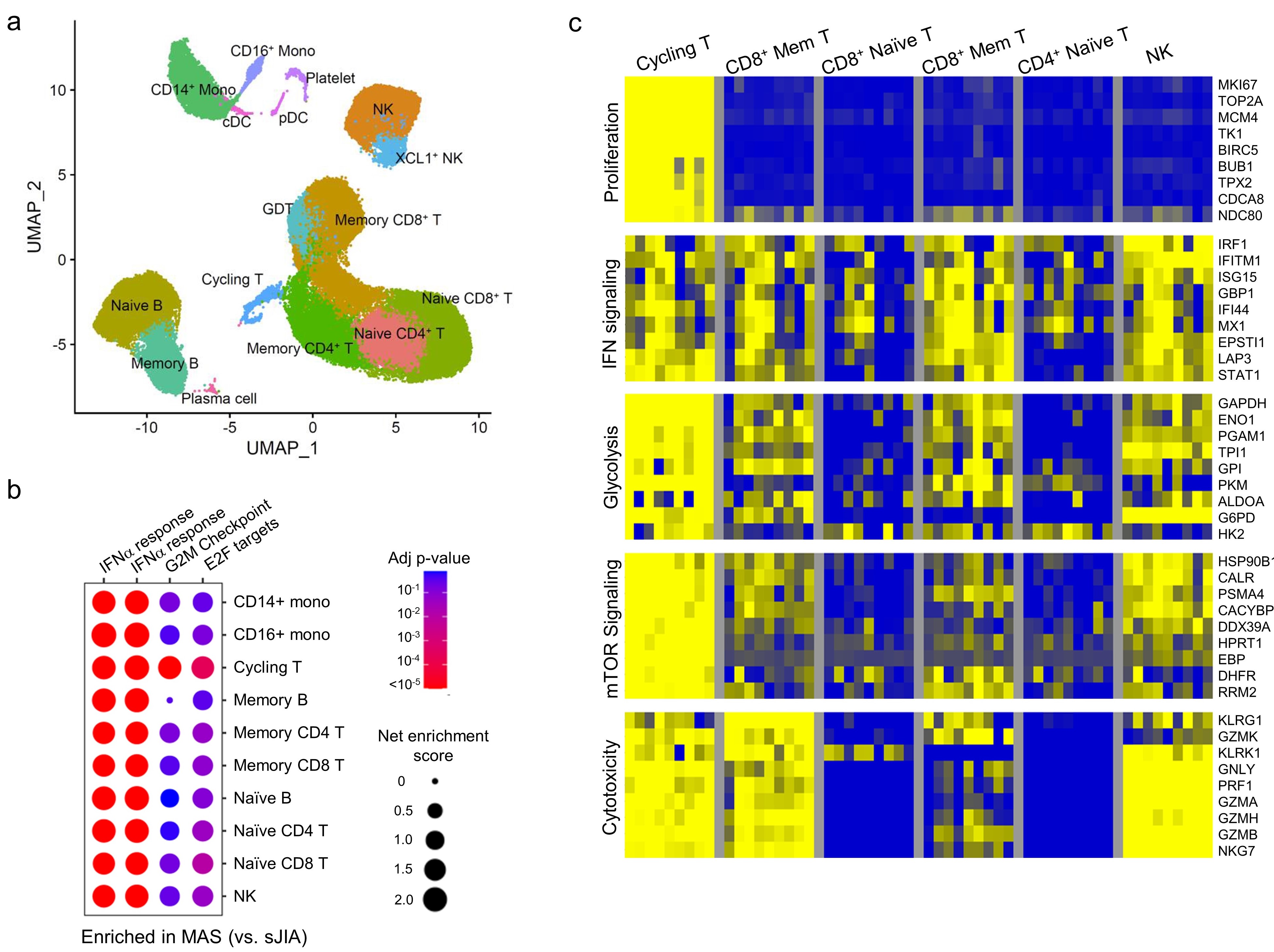 Figure 1. Single cell RNA-seq analysis of PBMCs from patients with sJIA with or without MAS. a) Uniform Manifold Approximation and Projection (UMAP) and annotation of immune cell populations. b) cluster plot of gene set enrichment analysis comparing the gene signatures of IFN-I, IFN-I and cell proliferation (G2M checkpoint and E2F targets). C) heatmap comparison of gene signatures across different lymphocyte populations in patients with sJIA-associated MAS.
Figure 1. Single cell RNA-seq analysis of PBMCs from patients with sJIA with or without MAS. a) Uniform Manifold Approximation and Projection (UMAP) and annotation of immune cell populations. b) cluster plot of gene set enrichment analysis comparing the gene signatures of IFN-I, IFN-I and cell proliferation (G2M checkpoint and E2F targets). C) heatmap comparison of gene signatures across different lymphocyte populations in patients with sJIA-associated MAS.
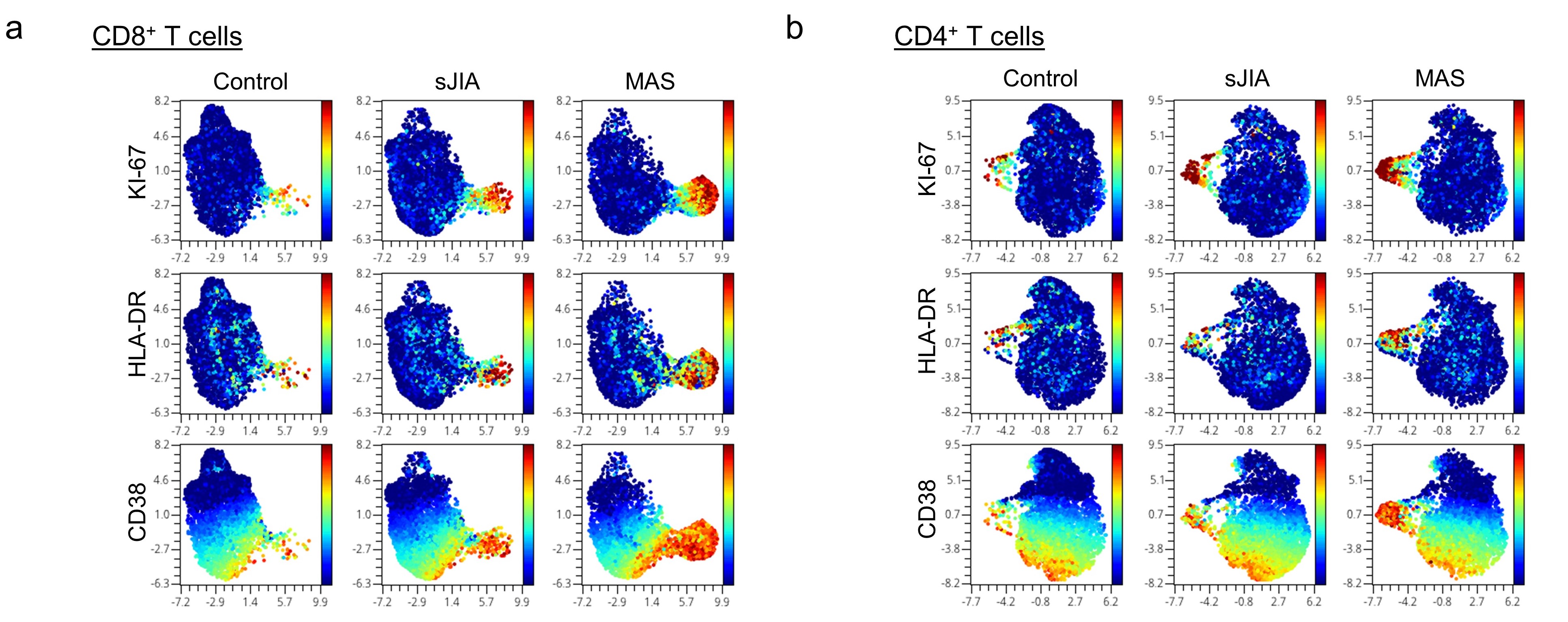 Figure 2. UMAP display of mass cytometry analysis comparing PBMCs from healthy controls and patients with sJIA with or without MAS. Color denotes the expression of KI-67, HLA-DR and CD38) on a) CD8+ T cells and b) CD4+ T cells.
Figure 2. UMAP display of mass cytometry analysis comparing PBMCs from healthy controls and patients with sJIA with or without MAS. Color denotes the expression of KI-67, HLA-DR and CD38) on a) CD8+ T cells and b) CD4+ T cells.
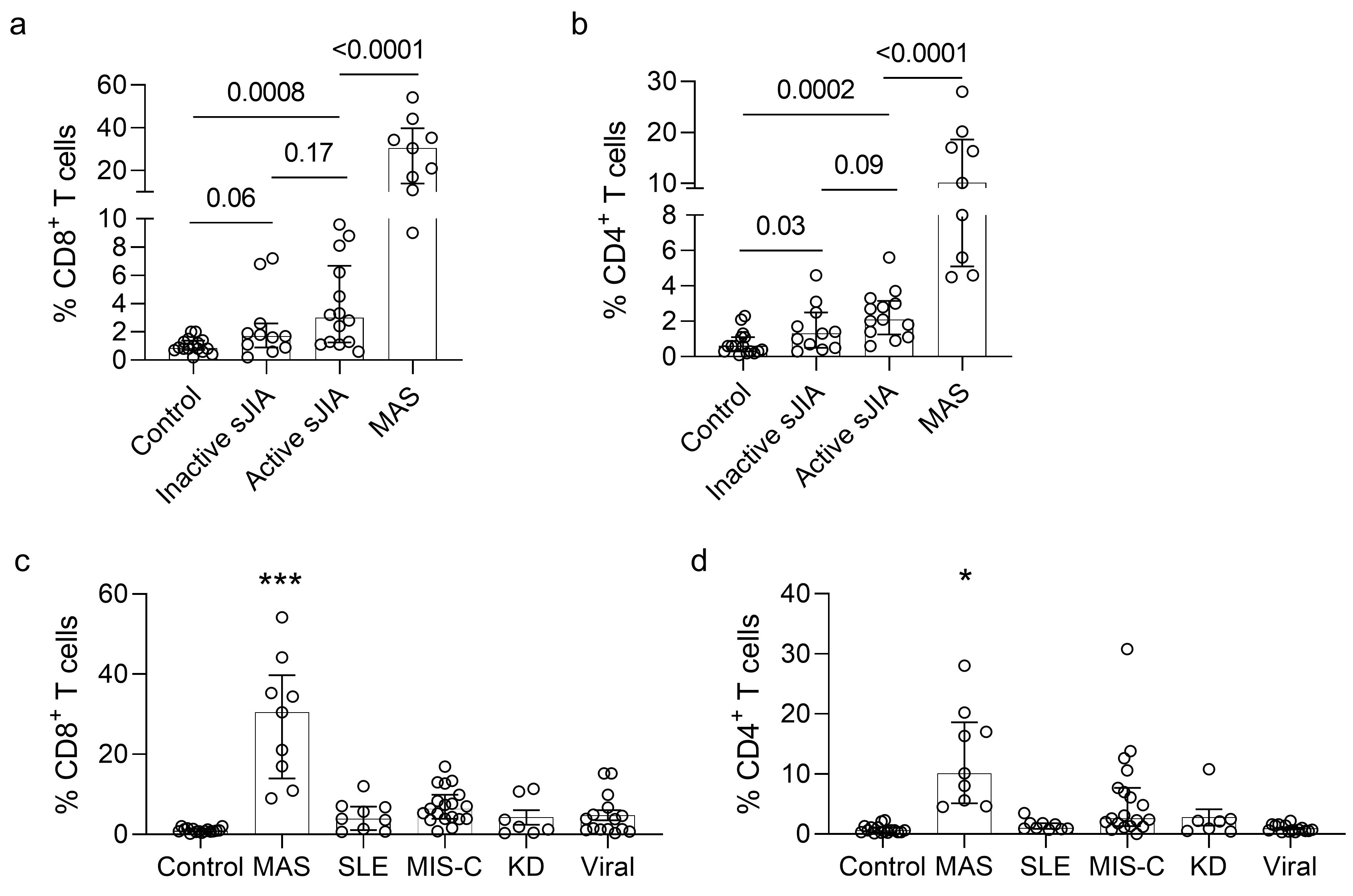 Figure 2. Quantification of cycling T cells in sJIA and other pediatric inflammatory diseases. a) Quantification of CD8+CD38+HLA-DR+ and b) CD4+CD38+HLA-DR+ T cells in healthy controls and patients with inactive sJIA, active sJIA without MAS, and active sJIA with MAS. c) Comparison of CD8+CD38+HLA-DR+ and d) CD4+CD38+HLA-DR+ T cells in healthy controls and patients with MAS, MIS-C, SLE, KD, and acute viral infections. * p < 0.05; *** p < 0.001
Figure 2. Quantification of cycling T cells in sJIA and other pediatric inflammatory diseases. a) Quantification of CD8+CD38+HLA-DR+ and b) CD4+CD38+HLA-DR+ T cells in healthy controls and patients with inactive sJIA, active sJIA without MAS, and active sJIA with MAS. c) Comparison of CD8+CD38+HLA-DR+ and d) CD4+CD38+HLA-DR+ T cells in healthy controls and patients with MAS, MIS-C, SLE, KD, and acute viral infections. * p < 0.05; *** p < 0.001
Disclosures: K. Brodeur, None; L. Chen, None; z. huang, None; Y. Du, None; H. Wobma, Immplacate Inc; M. Taylor, None; J. Chang, GlaxoSmithKline (GSK); M. Day-Lewis, None; F. Dedeoglu, Novartis; O. Halyabar, None; M. Lo, GlaxoSmithKlein(GSK); J. Newburger, None; M. Son, None; R. Sundel, None; P. Nigrovic, Bristol-Myers Squibb(BMS), Exo Therapeutics, Miach Orthopedics, Pfizer, Novartis, UpToDate, Cerecor, Brickell Bio; l. henderson, Sobi, Pfizer, Adaptive Biotechnologies, BMS; P. Lee, None.
Background/Purpose: Macrophage activation syndrome (MAS) is a complication of systemic juvenile idiopathic arthritis (sJIA) characterized by cytokine storm and overt immune cell activation. We aim to further understand the immunologic landscape of sJIA and MAS.
Methods: We profiled peripheral blood mononuclear cells (PBMCs) from patients with active sJIA with or without MAS at the time of sampling and age-matched controls using bulk RNA sequencing (RNA-seq), single-cell RNA-seq, and mass cytometry. All patients with sJIA met ILAR diagnostic criteria and MAS was defined according to the 2016 Classification criteria for MAS complicating sJIA. Findings from RNA-seq and mass cytometry were validated using additional samples from patients with sJIA as well as patients with other inflammatory diseases by flow cytometry and in vitro T cell stimulation studies.
Results: Gene set enrichment analysis of bulk RNA-seq of PBMCs from patients with MAS (n = 9) revealed strong expression of genes associated with type I interferon signaling and T cell proliferation in addition to the expected type II IFN (IFN-g) signature based on gene set enrichment analysis. These features were rarely seen in age-matched healthy controls (n = 18) or patients with sJIA without MAS (n = 15). Single-cell RNA-seq confirmed the presence of type I and type II IFN signatures on all major PBMC subsets in patients with MAS (n = 8) while the T cell proliferation signature was localized to a population of Ki67+ cycling T cells with enhanced expression of IFN-g and genes involved in glycolysis, mTOR signaling and cytotoxicity (Figure 1).
Mass cytometry revealed that the cycling T cell population comprised of predominantly CD8+CD38+HLA-DR+ T cells and fewer CD4+CD38+HLA-DR+ T cells (Figure 2). We found that an expansion of CD38+HLA-DR+ T cells beyond 9% of CD8+ T cells or 4% of CD4+ T cells distinguished MAS from active sJIA, while a milder increase was generally seen in patients with sJIA without MAS compared to healthy controls (Figure 3). The degree of CD8+CD38+HLA-DR+ T cell expansion was also significantly greater in MAS compared to multi-system inflammatory syndrome in children (MIS-C) associated with COVID-19 (n = 17), acute viral infections (n = 15), Kawasaki disease (n = 7) and systemic lupus erythematosus (n = 9). In addition, stimulation of isolated T cells from healthy donors with IFN-I, but not IFN-II, augmented the effects of IL-12, IL-15 and IL-18 to generate CD8+CD38+HLA-DR+ T cells in vitro, while Janus kinase inhibition by ruxolitinib mitigated the impact of IFN-I treatment.
Conclusion: Our multi-omic analysis further details the expansion of the recently described CD8+CD38+HLADR+ T cells in MAS associated with sJIA. Our findings demonstrate a synergistic role of IFN-I in the generation of these cycling T cells and provide a mechanistic link between viral infections and MAS that may be targetable by JAK inhibition.
 Figure 1. Single cell RNA-seq analysis of PBMCs from patients with sJIA with or without MAS. a) Uniform Manifold Approximation and Projection (UMAP) and annotation of immune cell populations. b) cluster plot of gene set enrichment analysis comparing the gene signatures of IFN-I, IFN-I and cell proliferation (G2M checkpoint and E2F targets). C) heatmap comparison of gene signatures across different lymphocyte populations in patients with sJIA-associated MAS.
Figure 1. Single cell RNA-seq analysis of PBMCs from patients with sJIA with or without MAS. a) Uniform Manifold Approximation and Projection (UMAP) and annotation of immune cell populations. b) cluster plot of gene set enrichment analysis comparing the gene signatures of IFN-I, IFN-I and cell proliferation (G2M checkpoint and E2F targets). C) heatmap comparison of gene signatures across different lymphocyte populations in patients with sJIA-associated MAS. Figure 2. UMAP display of mass cytometry analysis comparing PBMCs from healthy controls and patients with sJIA with or without MAS. Color denotes the expression of KI-67, HLA-DR and CD38) on a) CD8+ T cells and b) CD4+ T cells.
Figure 2. UMAP display of mass cytometry analysis comparing PBMCs from healthy controls and patients with sJIA with or without MAS. Color denotes the expression of KI-67, HLA-DR and CD38) on a) CD8+ T cells and b) CD4+ T cells.  Figure 2. Quantification of cycling T cells in sJIA and other pediatric inflammatory diseases. a) Quantification of CD8+CD38+HLA-DR+ and b) CD4+CD38+HLA-DR+ T cells in healthy controls and patients with inactive sJIA, active sJIA without MAS, and active sJIA with MAS. c) Comparison of CD8+CD38+HLA-DR+ and d) CD4+CD38+HLA-DR+ T cells in healthy controls and patients with MAS, MIS-C, SLE, KD, and acute viral infections. * p < 0.05; *** p < 0.001
Figure 2. Quantification of cycling T cells in sJIA and other pediatric inflammatory diseases. a) Quantification of CD8+CD38+HLA-DR+ and b) CD4+CD38+HLA-DR+ T cells in healthy controls and patients with inactive sJIA, active sJIA without MAS, and active sJIA with MAS. c) Comparison of CD8+CD38+HLA-DR+ and d) CD4+CD38+HLA-DR+ T cells in healthy controls and patients with MAS, MIS-C, SLE, KD, and acute viral infections. * p < 0.05; *** p < 0.001Disclosures: K. Brodeur, None; L. Chen, None; z. huang, None; Y. Du, None; H. Wobma, Immplacate Inc; M. Taylor, None; J. Chang, GlaxoSmithKline (GSK); M. Day-Lewis, None; F. Dedeoglu, Novartis; O. Halyabar, None; M. Lo, GlaxoSmithKlein(GSK); J. Newburger, None; M. Son, None; R. Sundel, None; P. Nigrovic, Bristol-Myers Squibb(BMS), Exo Therapeutics, Miach Orthopedics, Pfizer, Novartis, UpToDate, Cerecor, Brickell Bio; l. henderson, Sobi, Pfizer, Adaptive Biotechnologies, BMS; P. Lee, None.

