Back
Poster Session B
Systemic lupus erythematosus (SLE)
Session: (0629–0670) SLE – Etiology and Pathogenesis Poster
0662: Partial Correlations Network Models Show Th1, Th2 and Th17 Responses to Be Interlinked in Dermal Pathogenesis of Cutaneous Lupus Erythematous
Sunday, November 13, 2022
9:00 AM – 10:30 AM Eastern Time
Location: Virtual Poster Hall
- FC
Felix Chin, BS
University of Pennsylvania Perelman School of Medicine
Philadelphia, PA, United States
Abstract Poster Presenter(s)
Felix Chin1, Thomas Vazquez2, Josh Dan3, DeAnna Diaz4, Grant Sprow5, Jay Patel6, Nilesh Kodali7, Rui Feng8 and Victoria Werth9, 1University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, 2FIU Wertheim College of Medicine, Virginia Beach, VA, 3Philadelphia VAMC, Philadelphia, PA, USA and Department of Dermatology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, 4Philadelphia College of Medicine, Philadelphia, PA, 5Albert Einstein College of Medicine, Philadelphia, PA, 6Philadelphia VAMC, Philadelphia, PA, USA and Department of Dermatology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PN, 7New Jersey Medical School, Coppell, TX, 8University of Pennsylvania, Philadelphia, 9University of Pennsylvania and Corporal Michael J. Crescenz VA Medical Center, Philadelphia, PA
Background/Purpose: The immunopathogenesis of cutaneous lupus erythematous (CLE) is highly diverse and involves activity of many different cell types and pathways. This heterogeneity is believed to be largely responsible for unpredictable treatment response between patients. Here we used a correlation network model approach to map the relationships between different cell types and pathway proteins in CLE skin biopsies.
Methods: Here we profiled 44 CLE biopsies using multiplexed imaging mass cytometry to characterize the cell and pathway composition of CLE infiltrate. Biopsies were stained with two different panels of 37-metal conjugated antibodies that served as markers for different cell types, cytokines and pathway proteins. The relationships among cell and cytokine immune markers were modeled using Gaussian graphical model. The values were log-transformed and standardized prior to the analysis. The GGM algorithm identifies and displays only significant correlations between markers while leaving out non-significant ones. The algorithm then graphs the markers as a network and spatially arranges them by correlation strength so that the strongest associated markers are within closest proximity. This spatial orientation allows for distinction between direct and indirect associations of cell types or pathway proteins.
Results: Of our network consisting of 11 T-cell subtypes, GGM identified Th1, Th2 and Th17 to be inter-related with each other, forming a triangle. No other positive findings were found between any other cells, including our main cell network consisting of 11 common immune cell types. Of our cytokine/pathway network consisting of 16 different markers, GGM identified IL4, IL17 and IFN𝛾 to be among the strongest related markers in the network. Correlation testing confirmed remarkably strong associations between IL4, IL17 and IFN𝛾 (all 3 pairs were p< 1x10-13) as well as significant correlations (p< 1x10-6) amongst all possible permutations of the aforementioned GGM-significant markers. Furthermore, heatmap visualization of T-cell K-means clustering was also performed and revealed the majority of patients (n=27) to be Tem dominant (83% CD4 and 76% CD8), with a smaller subgroup (n=14) to being Tcm dominant (57% CD4 and 50% CD8).
Conclusion: These findings suggest that Th1, Th2 and Th17 co-occur together in the pathogenesis of CLE. Th1 and Th2 responses have traditionally been thought of as separate and antagonistic to each other. Our study challenges this dualism. In treating CLE patients with increased T-cell activity, it may be necessary to tackle both Th1 and Th2 along with Th17 responses together.
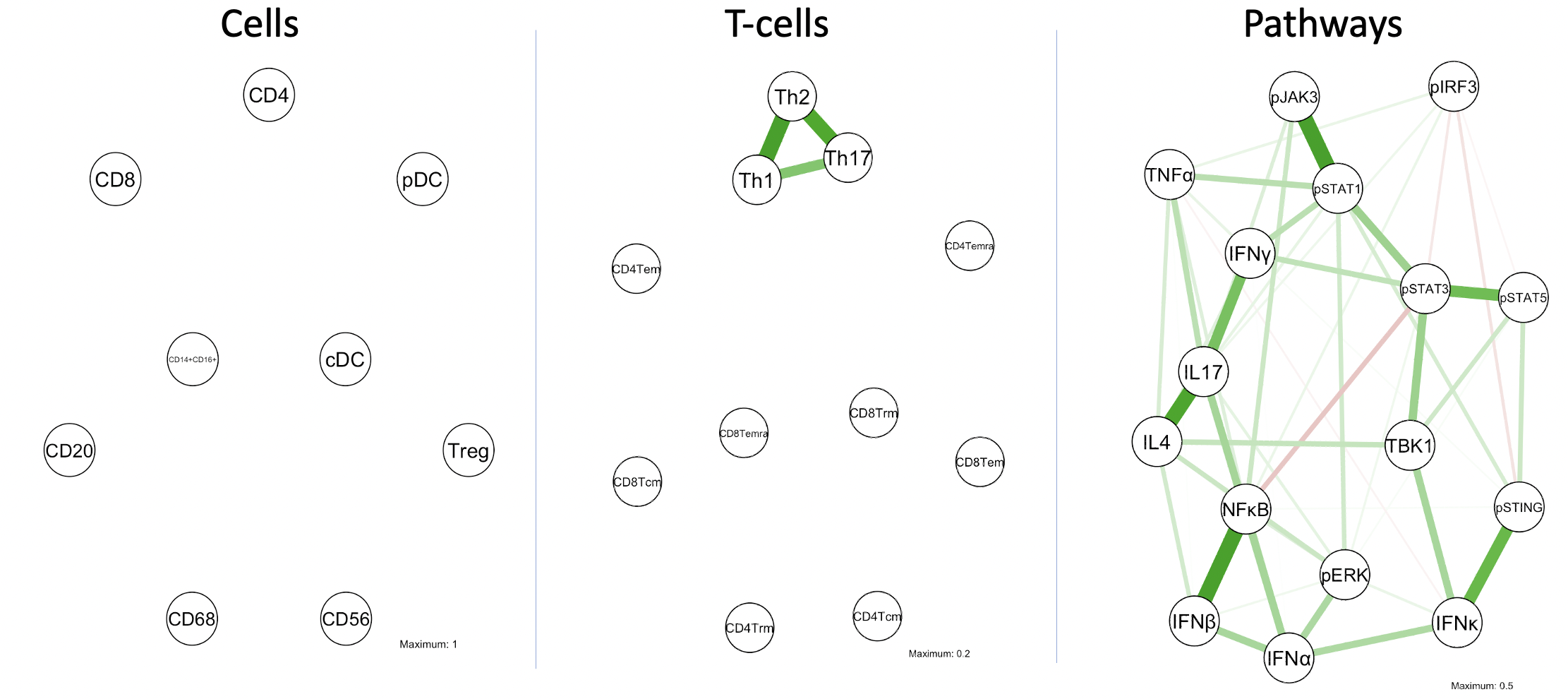 Fig 1. GGM networks of cells, T-cell subsets and pathway proteins. Lines indicate significant correlations between nodes. Green lines indicate positive association while red lines indicate a negative association. Line color intensity and spatial proximity between nodes is a reflection of correlation strength.
Fig 1. GGM networks of cells, T-cell subsets and pathway proteins. Lines indicate significant correlations between nodes. Green lines indicate positive association while red lines indicate a negative association. Line color intensity and spatial proximity between nodes is a reflection of correlation strength.
.jpeg) Table 1. Spearman's R correlation coefficient strengths between all possible pairs of cells or cytokines involved in Th1, Th2 and Th17 response.
Table 1. Spearman's R correlation coefficient strengths between all possible pairs of cells or cytokines involved in Th1, Th2 and Th17 response.
.jpeg) Fig 2. Linear relationship between IL4 and IL17 levels across 44 patients studied.
Fig 2. Linear relationship between IL4 and IL17 levels across 44 patients studied.
Disclosures: F. Chin, None; T. Vazquez, None; J. Dan, None; D. Diaz, None; G. Sprow, None; J. Patel, None; N. Kodali, None; R. Feng, None; V. Werth, AbbVie/Abbott, Amgen, AstraZeneca, Bristol Myers Squibb (BMS), Celgene, Eli Lilly, Genentech, GlaxoSmithKline (GSK), Janssen, Merck/MSD, Gilead, Novartis, Pfizer, Rome Pharmaceuticals, Horizon Therapeutics, Regeneron, argenx, CSL Behring, AnaptysBio, Biogen, Corbus, EMD Serono, Galderma, Nektar, Octapharma, Principia, Resolve, Sanofi, Syntimmune, Viela.
Background/Purpose: The immunopathogenesis of cutaneous lupus erythematous (CLE) is highly diverse and involves activity of many different cell types and pathways. This heterogeneity is believed to be largely responsible for unpredictable treatment response between patients. Here we used a correlation network model approach to map the relationships between different cell types and pathway proteins in CLE skin biopsies.
Methods: Here we profiled 44 CLE biopsies using multiplexed imaging mass cytometry to characterize the cell and pathway composition of CLE infiltrate. Biopsies were stained with two different panels of 37-metal conjugated antibodies that served as markers for different cell types, cytokines and pathway proteins. The relationships among cell and cytokine immune markers were modeled using Gaussian graphical model. The values were log-transformed and standardized prior to the analysis. The GGM algorithm identifies and displays only significant correlations between markers while leaving out non-significant ones. The algorithm then graphs the markers as a network and spatially arranges them by correlation strength so that the strongest associated markers are within closest proximity. This spatial orientation allows for distinction between direct and indirect associations of cell types or pathway proteins.
Results: Of our network consisting of 11 T-cell subtypes, GGM identified Th1, Th2 and Th17 to be inter-related with each other, forming a triangle. No other positive findings were found between any other cells, including our main cell network consisting of 11 common immune cell types. Of our cytokine/pathway network consisting of 16 different markers, GGM identified IL4, IL17 and IFN𝛾 to be among the strongest related markers in the network. Correlation testing confirmed remarkably strong associations between IL4, IL17 and IFN𝛾 (all 3 pairs were p< 1x10-13) as well as significant correlations (p< 1x10-6) amongst all possible permutations of the aforementioned GGM-significant markers. Furthermore, heatmap visualization of T-cell K-means clustering was also performed and revealed the majority of patients (n=27) to be Tem dominant (83% CD4 and 76% CD8), with a smaller subgroup (n=14) to being Tcm dominant (57% CD4 and 50% CD8).
Conclusion: These findings suggest that Th1, Th2 and Th17 co-occur together in the pathogenesis of CLE. Th1 and Th2 responses have traditionally been thought of as separate and antagonistic to each other. Our study challenges this dualism. In treating CLE patients with increased T-cell activity, it may be necessary to tackle both Th1 and Th2 along with Th17 responses together.
 Fig 1. GGM networks of cells, T-cell subsets and pathway proteins. Lines indicate significant correlations between nodes. Green lines indicate positive association while red lines indicate a negative association. Line color intensity and spatial proximity between nodes is a reflection of correlation strength.
Fig 1. GGM networks of cells, T-cell subsets and pathway proteins. Lines indicate significant correlations between nodes. Green lines indicate positive association while red lines indicate a negative association. Line color intensity and spatial proximity between nodes is a reflection of correlation strength. .jpeg) Table 1. Spearman's R correlation coefficient strengths between all possible pairs of cells or cytokines involved in Th1, Th2 and Th17 response.
Table 1. Spearman's R correlation coefficient strengths between all possible pairs of cells or cytokines involved in Th1, Th2 and Th17 response. .jpeg) Fig 2. Linear relationship between IL4 and IL17 levels across 44 patients studied.
Fig 2. Linear relationship between IL4 and IL17 levels across 44 patients studied.Disclosures: F. Chin, None; T. Vazquez, None; J. Dan, None; D. Diaz, None; G. Sprow, None; J. Patel, None; N. Kodali, None; R. Feng, None; V. Werth, AbbVie/Abbott, Amgen, AstraZeneca, Bristol Myers Squibb (BMS), Celgene, Eli Lilly, Genentech, GlaxoSmithKline (GSK), Janssen, Merck/MSD, Gilead, Novartis, Pfizer, Rome Pharmaceuticals, Horizon Therapeutics, Regeneron, argenx, CSL Behring, AnaptysBio, Biogen, Corbus, EMD Serono, Galderma, Nektar, Octapharma, Principia, Resolve, Sanofi, Syntimmune, Viela.

