Back
Poster Session B
Rheumatoid arthritis (RA)
Session: (0589–0628) RA – Etiology and Pathogenesis Poster
0610: Patient-derived Organoids Reveal Transcriptional Regulation of Synovial Lining Fibroblast Differentiation
Sunday, November 13, 2022
9:00 AM – 10:30 AM Eastern Time
Location: Virtual Poster Hall
- SP
Sonia Presti, BS
Brigham and Women's Hospital
Boston, MA, United States
Abstract Poster Presenter(s)
Sonia Presti1, Gerald FM Watts1, Zhu Zhu1, Kartik Bhamidipati1, Junning Case1, Yuhong Li1, Teri Bowman1, Suppawat Kongthong1, ilya Korsunsky1, Michael Brenner2 and Kevin Wei3, 1Brigham and Women's Hospital, Boston, MA, 2Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 3Brigham and Women's Hospital and Harvard Medical School, Boston, MA
Background/Purpose: Synovial lining fibroblasts secrete factors into the joint cavity that promote joint lubrication1. In inflammatory arthritis, sublining fibroblasts undergo marked expansion while lining fibroblasts are lost, leading to joint damage2,3,4. While we previously reported the role of Notch signaling in driving sublining fibroblast differentiation, molecular signals driving lining fibroblasts remain unknown.
Methods:
Conclusion: Our studies show that GSK3i induces a phenotypic and transcriptional switch towards synovial lining-like fibroblasts. Further investigations are underway to identify transcription factors necessary for lining fibroblast differentiation. Identification of transcriptional regulators will shed light on the molecular mechanism behind fibroblast heterogeneity and provide therapeutic targets for restoring synovial lining function in inflammatory arthritis.
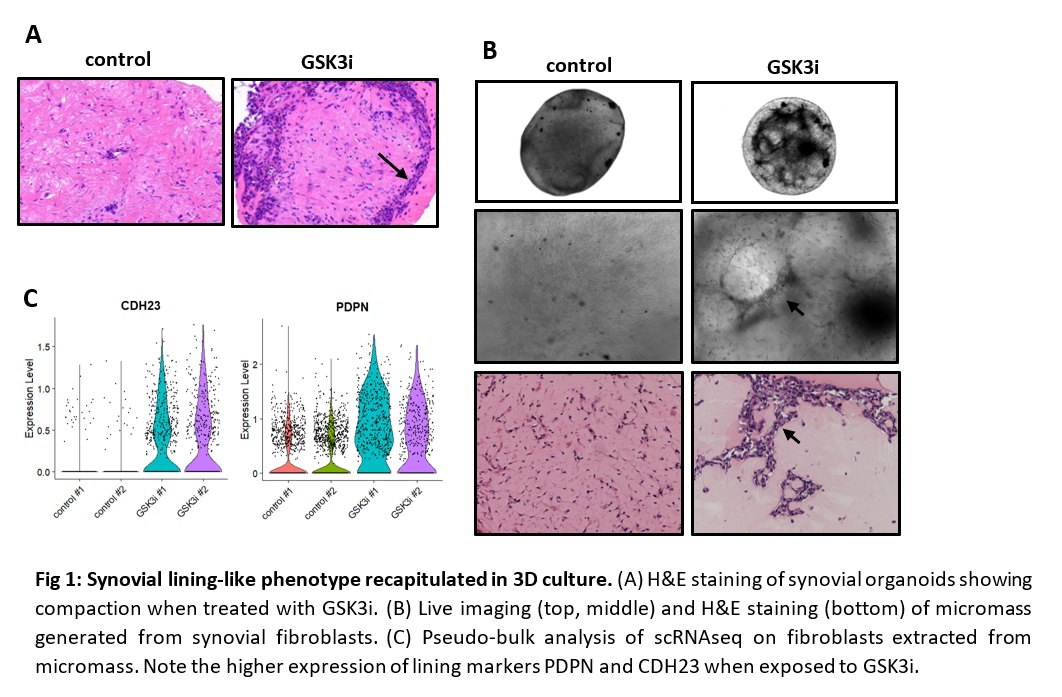 Fig 1: Synovial lining-like phenotype recapitulated in 3D culture. (A) H&E staining of synovial organoids showing compaction when treated with GSK3i. (B) Live imaging (top, middle) and H&E staining (bottom) of micromass generated from synovial fibroblasts. (C) Pseudo-bulk analysis of scRNAseq on fibroblasts extracted from micromass. Note the higher expression of lining markers PDPN and CDH23 when exposed to GSK3i.
Fig 1: Synovial lining-like phenotype recapitulated in 3D culture. (A) H&E staining of synovial organoids showing compaction when treated with GSK3i. (B) Live imaging (top, middle) and H&E staining (bottom) of micromass generated from synovial fibroblasts. (C) Pseudo-bulk analysis of scRNAseq on fibroblasts extracted from micromass. Note the higher expression of lining markers PDPN and CDH23 when exposed to GSK3i.
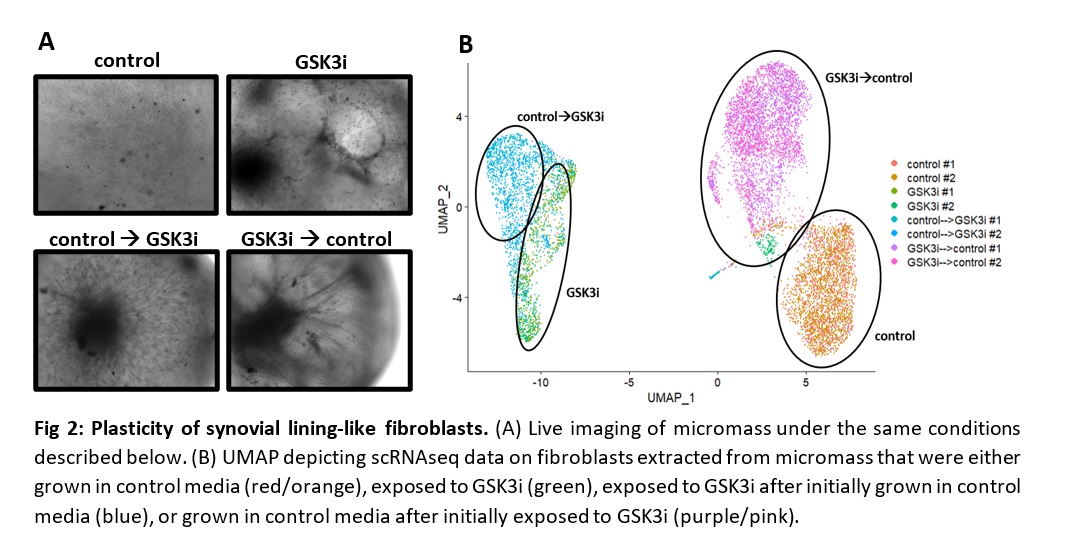 Fig 2: Plasticity of synovial lining-like fibroblasts. (A) Live imaging of micromass under the same conditions described below. (B) UMAP depicting scRNAseq data on fibroblasts extracted from micromass that were either grown in control media (red/orange), exposed to GSK3i (green), exposed to GSK3i after initially grown in control media (blue), or grown in control media after initially exposed to GSK3i (purple/pink).
Fig 2: Plasticity of synovial lining-like fibroblasts. (A) Live imaging of micromass under the same conditions described below. (B) UMAP depicting scRNAseq data on fibroblasts extracted from micromass that were either grown in control media (red/orange), exposed to GSK3i (green), exposed to GSK3i after initially grown in control media (blue), or grown in control media after initially exposed to GSK3i (purple/pink).
Disclosures: S. Presti, None; G. Watts, None; Z. Zhu, None; K. Bhamidipati, None; J. Case, None; Y. Li, None; T. Bowman, None; S. Kongthong, None; i. Korsunsky, Mestag Therapeutics Ltd.; M. Brenner, GSK, 4FO Ventures, Mestag Therapeutics; K. Wei, Gilead sciences, Mestag, nanoString, 10X Genomics.
Background/Purpose: Synovial lining fibroblasts secrete factors into the joint cavity that promote joint lubrication1. In inflammatory arthritis, sublining fibroblasts undergo marked expansion while lining fibroblasts are lost, leading to joint damage2,3,4. While we previously reported the role of Notch signaling in driving sublining fibroblast differentiation, molecular signals driving lining fibroblasts remain unknown.
Methods:
- Patient-derived organoids. We developed an en-block, patient-derived organoid to culture synovial tissue that retains fibroblast heterogeneity ex vivo.
- Micromass model. We resuspended cultured synovial fibroblasts in Matrigel to create an in vitro 3D model.
- Phenotypic analyses. We employed live imaging of organoids, histology, and single-cell RNA sequencing (scRNAseq) to characterize fibroblast phenotypes.
Conclusion: Our studies show that GSK3i induces a phenotypic and transcriptional switch towards synovial lining-like fibroblasts. Further investigations are underway to identify transcription factors necessary for lining fibroblast differentiation. Identification of transcriptional regulators will shed light on the molecular mechanism behind fibroblast heterogeneity and provide therapeutic targets for restoring synovial lining function in inflammatory arthritis.
- Bartok, B. & Firestein, G. S. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol. Rev. 233, 233–255 (2010).
- Mizoguchi, F. et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat. Commun. 9, 789 (2018).
- Zhang, F. et al. Defining Inflammatory Cell States in Rheumatoid Arthritis Joint Synovial Tissues by Integrating Single-cell Transcriptomics and Mass Cytometry. doi:10.1101/351130.
- Wei, K. et al. Notch signaling drives synovial fibroblast identity and arthritis pathology. Nature 582, 259–264 (2020).
- Zhang, F. et al. Cellular deconstruction of inflamed synovium defines diverse inflammatory phenotypes in rheumatoid arthritis. bioRxiv 2022.02.25.481990 (2022) doi:10.1101/2022.02.25.481990.
 Fig 1: Synovial lining-like phenotype recapitulated in 3D culture. (A) H&E staining of synovial organoids showing compaction when treated with GSK3i. (B) Live imaging (top, middle) and H&E staining (bottom) of micromass generated from synovial fibroblasts. (C) Pseudo-bulk analysis of scRNAseq on fibroblasts extracted from micromass. Note the higher expression of lining markers PDPN and CDH23 when exposed to GSK3i.
Fig 1: Synovial lining-like phenotype recapitulated in 3D culture. (A) H&E staining of synovial organoids showing compaction when treated with GSK3i. (B) Live imaging (top, middle) and H&E staining (bottom) of micromass generated from synovial fibroblasts. (C) Pseudo-bulk analysis of scRNAseq on fibroblasts extracted from micromass. Note the higher expression of lining markers PDPN and CDH23 when exposed to GSK3i. Fig 2: Plasticity of synovial lining-like fibroblasts. (A) Live imaging of micromass under the same conditions described below. (B) UMAP depicting scRNAseq data on fibroblasts extracted from micromass that were either grown in control media (red/orange), exposed to GSK3i (green), exposed to GSK3i after initially grown in control media (blue), or grown in control media after initially exposed to GSK3i (purple/pink).
Fig 2: Plasticity of synovial lining-like fibroblasts. (A) Live imaging of micromass under the same conditions described below. (B) UMAP depicting scRNAseq data on fibroblasts extracted from micromass that were either grown in control media (red/orange), exposed to GSK3i (green), exposed to GSK3i after initially grown in control media (blue), or grown in control media after initially exposed to GSK3i (purple/pink).Disclosures: S. Presti, None; G. Watts, None; Z. Zhu, None; K. Bhamidipati, None; J. Case, None; Y. Li, None; T. Bowman, None; S. Kongthong, None; i. Korsunsky, Mestag Therapeutics Ltd.; M. Brenner, GSK, 4FO Ventures, Mestag Therapeutics; K. Wei, Gilead sciences, Mestag, nanoString, 10X Genomics.

