Back
Poster Session B
Rheumatoid arthritis (RA)
Session: (0883–0912) RA – Diagnosis, Manifestations, and Outcomes Poster II
0905: Predicting Disease Activity in Rheumatoid Arthritis Patients Using Machine Learning: Data from the ACR’s RISE Registry
Sunday, November 13, 2022
9:00 AM – 10:30 AM Eastern Time
Location: Virtual Poster Hall
- ZI
Zara Izadi, PhD, PharmD
University of California San Francisco
San Francisco, CA, United States
Abstract Poster Presenter(s)
Eren-Ajani Tshimanga1, Milena Gianfrancesco2, Stefanos Giampanis3, Jing Li2, Emma Kersey4, Jinoos Yazdany5, Beau Norgeot6, Gabriela Schmajuk7 and Zara Izadi4, 1University of California, Berkeley, Berkeley, CA, 2University of California, San Francisco, San Francisco, CA, 3Anthem, San Francisco, 4University of California San Francisco, San Francisco, CA, 5UCSF, San Francisco, CA, 6Anthem, Oakland, CA, 7UCSF / SFVA, San Francisco, CA
Background/Purpose: Predicting the trajectory of disease in individuals with RA is difficult, with numerous factors influencing whether a patient may experience higher or lower disease activity at their next visit. Machine learning (ML) models that leverage variables available in electronic health records (EHRs) from a large number of patients have been used in many medical contexts to make predictions about the future. We assessed the ability of various ML models to predict disease activity in individuals with RA at future clinical visits using information from the current visit.
Methods: We used data through March 2021 from the ACR's RISE registry, a large, national EHR-based registry. We included individuals with ≥ 2 RA diagnoses (ICD-9: 714.0) ≥ 30 days apart, and ≥ 2 recorded clinical disease activity index (CDAI) scores ≥ 3 months apart. Disease activity was categorized into four levels (remission, low, moderate, and high activity) based on accepted score ranges. Data features used as predictors included age, sex, race, ethnicity, smoking, obesity, medications, and the previous visit's CDAI score. Missing data were imputed using last observation carried forward. Variables based on patient ZIP codes from the AHRQ SDOH Database were selected using LASSO and included in the models (total weighted population, per capita income, median home value, and median income of civilian population). The data was split into 80:20 train-test sets. We determined the performance of extreme gradient boosted trees (XGBoost), random forest (RF), and logistic regression (LR) models using sensitivity, specificity, and F-1 score, using 5-fold cross-validation. All models were compared to a baseline prediction model based only on the CDAI score from the previous visit.
Results: A total of 39,155 patients were included in the analysis. The mean (SD) age was 63.7 (13.5) years and 77.6% were female. The mean (SD) visits per patient was 3.9 (1.6). We observed that most patients' disease activity varied over time – 60.2% of patients experienced a change in CDAI score category at least once. The sensitivity, specificity, and F1-score of the baseline prediction (based only on the last observed CDAI score) vs. the best ML model (XGBoost) were very similar (0.8264, 0.9398, and 0.8234 vs. 0.8237, 0.9390, and 0.8209, respectively) (Table 2). Using XGBoost, the top three variables of importance that were identified were the previous CDAI category, previous raw CDAI score, and a binary feature indicating if the CDAI value was imputed.
Conclusion: ML models may be helpful in predicting RA disease activity in the future, although the performance of our best ML model was very similar to a standard baseline model in predicting CDAI score at a patient's next visit. Future studies may benefit from utilizing time-varying models such as "long short-term memory", which better capture the longitudinal nature of EHR data. These tools in conjunction with clinician judgments may allow for early action in managing disease activity for patients with RA.
Disclaimer: Data collection was supported by the ACR's RISE Registry. The views expressed represent those of the authors, not necessarily those of ACR.
.jpg) Characteristics of included individuals with rheumatoid arthritis in RISE at the first visit in the study period.
Characteristics of included individuals with rheumatoid arthritis in RISE at the first visit in the study period.
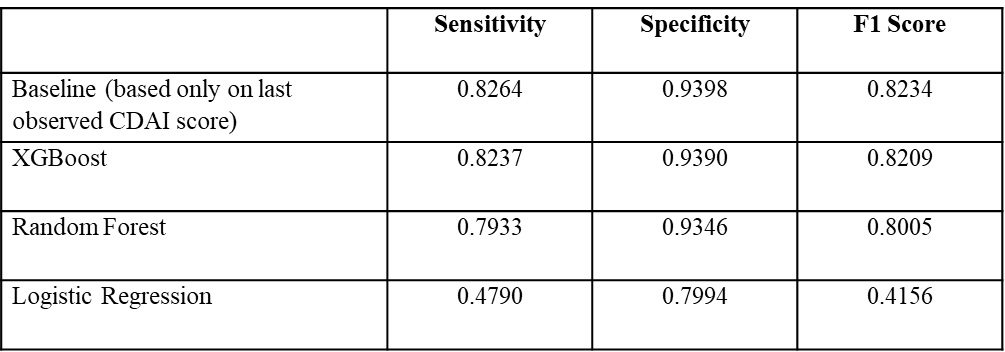 Model performance in the test set.
Model performance in the test set.
Disclosures: E. Tshimanga, None; M. Gianfrancesco, Pfizer; S. Giampanis, Anthem, Apple; J. Li, None; E. Kersey, None; J. Yazdany, AstraZeneca, Gilead, Bristol-Myers Squibb(BMS), Aurinia, Astra Zeneca, Pfizer; B. Norgeot, Anthem; G. Schmajuk, None; Z. Izadi, None.
Background/Purpose: Predicting the trajectory of disease in individuals with RA is difficult, with numerous factors influencing whether a patient may experience higher or lower disease activity at their next visit. Machine learning (ML) models that leverage variables available in electronic health records (EHRs) from a large number of patients have been used in many medical contexts to make predictions about the future. We assessed the ability of various ML models to predict disease activity in individuals with RA at future clinical visits using information from the current visit.
Methods: We used data through March 2021 from the ACR's RISE registry, a large, national EHR-based registry. We included individuals with ≥ 2 RA diagnoses (ICD-9: 714.0) ≥ 30 days apart, and ≥ 2 recorded clinical disease activity index (CDAI) scores ≥ 3 months apart. Disease activity was categorized into four levels (remission, low, moderate, and high activity) based on accepted score ranges. Data features used as predictors included age, sex, race, ethnicity, smoking, obesity, medications, and the previous visit's CDAI score. Missing data were imputed using last observation carried forward. Variables based on patient ZIP codes from the AHRQ SDOH Database were selected using LASSO and included in the models (total weighted population, per capita income, median home value, and median income of civilian population). The data was split into 80:20 train-test sets. We determined the performance of extreme gradient boosted trees (XGBoost), random forest (RF), and logistic regression (LR) models using sensitivity, specificity, and F-1 score, using 5-fold cross-validation. All models were compared to a baseline prediction model based only on the CDAI score from the previous visit.
Results: A total of 39,155 patients were included in the analysis. The mean (SD) age was 63.7 (13.5) years and 77.6% were female. The mean (SD) visits per patient was 3.9 (1.6). We observed that most patients' disease activity varied over time – 60.2% of patients experienced a change in CDAI score category at least once. The sensitivity, specificity, and F1-score of the baseline prediction (based only on the last observed CDAI score) vs. the best ML model (XGBoost) were very similar (0.8264, 0.9398, and 0.8234 vs. 0.8237, 0.9390, and 0.8209, respectively) (Table 2). Using XGBoost, the top three variables of importance that were identified were the previous CDAI category, previous raw CDAI score, and a binary feature indicating if the CDAI value was imputed.
Conclusion: ML models may be helpful in predicting RA disease activity in the future, although the performance of our best ML model was very similar to a standard baseline model in predicting CDAI score at a patient's next visit. Future studies may benefit from utilizing time-varying models such as "long short-term memory", which better capture the longitudinal nature of EHR data. These tools in conjunction with clinician judgments may allow for early action in managing disease activity for patients with RA.
Disclaimer: Data collection was supported by the ACR's RISE Registry. The views expressed represent those of the authors, not necessarily those of ACR.
.jpg) Characteristics of included individuals with rheumatoid arthritis in RISE at the first visit in the study period.
Characteristics of included individuals with rheumatoid arthritis in RISE at the first visit in the study period.  Model performance in the test set.
Model performance in the test set. Disclosures: E. Tshimanga, None; M. Gianfrancesco, Pfizer; S. Giampanis, Anthem, Apple; J. Li, None; E. Kersey, None; J. Yazdany, AstraZeneca, Gilead, Bristol-Myers Squibb(BMS), Aurinia, Astra Zeneca, Pfizer; B. Norgeot, Anthem; G. Schmajuk, None; Z. Izadi, None.

