Back
Abstract Session
Session: Abstracts: Fibromyalgia and Other Clinical Pain Syndromes (2233–2236)
2233: Machine Learning Uncovers Novel mRNAs Expressed in Fibromyalgia
Monday, November 14, 2022
4:30 PM – 4:40 PM Eastern Time
Location: Ballroom AB
- GS
Geoffrey Stephens, PhD
Exagen, Inc.
Vista, CA, United States
Presenting Author(s)
Lena Kolb1, Mark Rudolph1, Alan Kivitz2, Scott Rey1, Roberta Alexander1, Anja Kammesheidt3 and Geoffrey Stephens1, 1Exagen, Inc., Vista, CA, 2Department of Rheumatology, Altoona Center for Clinical Research, Duncansville, PA, 3self, Laguna Beach, CA
Background/Purpose: Fibromyalgia is a debilitating pain condition that affects roughly 12 million people in the United States. Although preliminary diagnostic criteria have been established by the ACR, the diffuse nature of the symptomology often precludes a definitive diagnosis. Here, we have identified a potential combination of biomarkers to enable a more objective diagnostic approach for fibromyalgia patients. To that end, a multi-center study was conducted to collect blood samples from diverse groups of patients whose symptomatology is often similar to fibromyalgia and whole transcriptome sequencing was performed.
Methods: Subjects: Samples from 26 individuals with fibromyalgia, 14 with systemic lupus erythematosus (SLE), 11 with rheumatoid arthritis (RA), and 14 normal donors (NHV) were analyzed. All patients satisfied the ACR classification criteria for their respective diseases.
Sequencing: RNA was isolated from whole blood collected in PAX gene tubes from a total of 65 individuals enrolled from five sites. After determination of the quality/quantity of the isolated total RNA, reverse transcription was used to generate cDNA to which adapters were ligated. High throughput sequencing was performed at a minimum depth of 25 million per sequence using a paired read approach.
Analysis: Raw data in the form of FASTQ files was mapped to the current version of the reference human genome (GRCh38) using Rsubread. Transcript counts were normalized, fold changes were calculated, and hierarchical clustering was performed using the R package DESeq2. Multiple machine learning algorithms were trained on a subset of the normalized count data, which was further validated using a separate test/holdout data set. The light gradient boosting model was subsequently determined to generate the most reliable predictions of disease benchmarked against ACR criteria. An analysis of significant pathways associated with the identified fibromyalgia specific gene set was performed using DAVID (https://david.ncifcrf.gov).
Results: Using a combination of traditional informatics and machine learning approaches, a distinct set of transcripts were identified in RNA isolated from the whole blood of patients with fibromyalgia, other conditions, and NHV (Figure 1). Fibromyalgia patients did not show elevated expression of type 1 IFN regulated genes, and analysis of pathway relationships showed an enrichment of genes involved in neuroactive peptide signaling, cognition, neuronal migration fibromyalgia. A machine learning approach revealed that the gene signature had significant potential value in facilitating fibromyalgia diagnosis with a detection sensitivity of 78% and a specificity of 93% (AUC of 0.96).
Conclusion: Next generation sequencing of RNA isolated from whole blood samples identified several genes that were differentially regulated in fibromyalgia patients versus those with other rheumatologic conditions. Using these data in the development of diagnostic algorithms may be a promising approach in distinguishing among diseases that have overlapping symptomology with fibromyalgia. Future studies using an orthogonal technology will be used to further validate the utility of the gene signature presented herein.
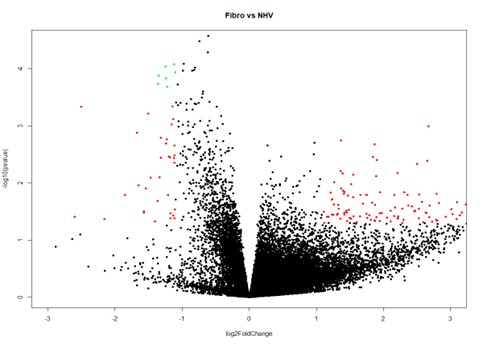 Figure 1. Volcano plot illustrating RNA transcripts differentially expressed (log2-fold changes) between 14 normal health donors (NHVs; x-axis) and 26 fibromyalgia (fibro; y-axis) patients. Red colored dots indicate transcripts that were differentially expressed with p-values < 0.05, and green dots indicate those with p-adjusted values of < 0.1.
Figure 1. Volcano plot illustrating RNA transcripts differentially expressed (log2-fold changes) between 14 normal health donors (NHVs; x-axis) and 26 fibromyalgia (fibro; y-axis) patients. Red colored dots indicate transcripts that were differentially expressed with p-values < 0.05, and green dots indicate those with p-adjusted values of < 0.1.
Disclosures: L. Kolb, Exagen; M. Rudolph, Exagen, Inc; A. Kivitz, Amgen, Boehringer-Ingelheim, Janssen, Gilead, GlaxoSmithKlein (GSK), Novartis, Pfizer, Sanofi, Flexion, Eli Lilly, Genentech, UCB, AbbVie, Merck, ECOR1 CAPITAL, LLC, Chemocentryx, Regenerson, Grunenthal, Bendcare, Horizon; S. Rey, Exagen Inc.; R. Alexander, Exagen Inc.; A. Kammesheidt, Exagen Inc.; G. Stephens, Exagen Inc.
Background/Purpose: Fibromyalgia is a debilitating pain condition that affects roughly 12 million people in the United States. Although preliminary diagnostic criteria have been established by the ACR, the diffuse nature of the symptomology often precludes a definitive diagnosis. Here, we have identified a potential combination of biomarkers to enable a more objective diagnostic approach for fibromyalgia patients. To that end, a multi-center study was conducted to collect blood samples from diverse groups of patients whose symptomatology is often similar to fibromyalgia and whole transcriptome sequencing was performed.
Methods: Subjects: Samples from 26 individuals with fibromyalgia, 14 with systemic lupus erythematosus (SLE), 11 with rheumatoid arthritis (RA), and 14 normal donors (NHV) were analyzed. All patients satisfied the ACR classification criteria for their respective diseases.
Sequencing: RNA was isolated from whole blood collected in PAX gene tubes from a total of 65 individuals enrolled from five sites. After determination of the quality/quantity of the isolated total RNA, reverse transcription was used to generate cDNA to which adapters were ligated. High throughput sequencing was performed at a minimum depth of 25 million per sequence using a paired read approach.
Analysis: Raw data in the form of FASTQ files was mapped to the current version of the reference human genome (GRCh38) using Rsubread. Transcript counts were normalized, fold changes were calculated, and hierarchical clustering was performed using the R package DESeq2. Multiple machine learning algorithms were trained on a subset of the normalized count data, which was further validated using a separate test/holdout data set. The light gradient boosting model was subsequently determined to generate the most reliable predictions of disease benchmarked against ACR criteria. An analysis of significant pathways associated with the identified fibromyalgia specific gene set was performed using DAVID (https://david.ncifcrf.gov).
Results: Using a combination of traditional informatics and machine learning approaches, a distinct set of transcripts were identified in RNA isolated from the whole blood of patients with fibromyalgia, other conditions, and NHV (Figure 1). Fibromyalgia patients did not show elevated expression of type 1 IFN regulated genes, and analysis of pathway relationships showed an enrichment of genes involved in neuroactive peptide signaling, cognition, neuronal migration fibromyalgia. A machine learning approach revealed that the gene signature had significant potential value in facilitating fibromyalgia diagnosis with a detection sensitivity of 78% and a specificity of 93% (AUC of 0.96).
Conclusion: Next generation sequencing of RNA isolated from whole blood samples identified several genes that were differentially regulated in fibromyalgia patients versus those with other rheumatologic conditions. Using these data in the development of diagnostic algorithms may be a promising approach in distinguishing among diseases that have overlapping symptomology with fibromyalgia. Future studies using an orthogonal technology will be used to further validate the utility of the gene signature presented herein.
 Figure 1. Volcano plot illustrating RNA transcripts differentially expressed (log2-fold changes) between 14 normal health donors (NHVs; x-axis) and 26 fibromyalgia (fibro; y-axis) patients. Red colored dots indicate transcripts that were differentially expressed with p-values < 0.05, and green dots indicate those with p-adjusted values of < 0.1.
Figure 1. Volcano plot illustrating RNA transcripts differentially expressed (log2-fold changes) between 14 normal health donors (NHVs; x-axis) and 26 fibromyalgia (fibro; y-axis) patients. Red colored dots indicate transcripts that were differentially expressed with p-values < 0.05, and green dots indicate those with p-adjusted values of < 0.1.Disclosures: L. Kolb, Exagen; M. Rudolph, Exagen, Inc; A. Kivitz, Amgen, Boehringer-Ingelheim, Janssen, Gilead, GlaxoSmithKlein (GSK), Novartis, Pfizer, Sanofi, Flexion, Eli Lilly, Genentech, UCB, AbbVie, Merck, ECOR1 CAPITAL, LLC, Chemocentryx, Regenerson, Grunenthal, Bendcare, Horizon; S. Rey, Exagen Inc.; R. Alexander, Exagen Inc.; A. Kammesheidt, Exagen Inc.; G. Stephens, Exagen Inc.

