Back
Poster Session B
Systemic lupus erythematosus (SLE)
Session: (0629–0670) SLE – Etiology and Pathogenesis Poster
0655: Proteomic Analysis of Histological Lesions Lupus Nephritis Identifies an Inflammatory Signature of Fibrous Crescents
Sunday, November 13, 2022
9:00 AM – 10:30 AM Eastern Time
Location: Virtual Poster Hall
- AC
Alessandra Ida Celia, MD
Johns Hopkins University
Baltimore, MD, United States
Abstract Poster Presenter(s)
Alessandra Ida Celia1, Jeffery Hodgin2, Avi Rosenberg3, Laurence S Magder4, Jill Buyon5, Betty Diamond6, Judith James7, William Apruzzese8, Paride Fenaroli9, Derek Fine1, Jose Monroy-Trujillo1, Mohamed G. Atta1, Peter izmirly10, Michael Belmont5, Anne Davidson6, Daniel W. Goldman11, the Accelerating Medicines Partnership (AMP) RA/SLE12, Michelle Petri11 and Andrea Fava1, 1Johns Hopkins University, Baltimore, MD, 2University of Michigan, Ann Arbor, MI, 3Johns Hopkins University, Ballwin, MO, 4University of Maryland, Department of Epidemiology and Public Health, Baltimore, MD, 5NYU Grossman School of Medicine, New York, NY, 6Feinstein Institutes for Medical Research, Manhasset, NY, 7Oklahoma Medical Research Foundation, Oklahoma City, OK, 8Brigham and Women's Hospital, Boston, MA, 9Università degli Studi di Parma, Parma, Emilia-Romagna, Italy, 10NYU Long Island School of Medicine, New York, NY, 11Johns Hopkins University School of Medicine, Division of Rheumatology, Baltimore, MD, 12Multiple Insitutions
Background/Purpose: We employed urine proteomics to define the molecular signatures associated with the histological features quantified by the NIH activity and chronicity indices.
Methods: Glomerular and interstitial lesions in lupus nephritis were quantified (scored 0-3) based on the revised 2018 International Society of Nephrology/Renal Pathology Society (ISN/RPS) classification for lupus nephritis and the modified NIH scoring system by a central renal pathologist (JH). Urinary proteins (1200 biomarkers, RayBiotech Kiloplex) were quantified in urine samples collected on the day of (73%) or within 3 weeks of (27%) the diagnostic kidney biopsy. Proteomic signatures of each lesion were defined based on Spearman correlations of each urine protein with each pathologic lesion.
Results: Ninety-one biopsies were included: 32 (35%) with pure proliferative LN, 33 (36%) with pure membranous LN, and 26 (29%) with mixed LN. The 5 most correlated urinary proteins and each pathologic feature are summarized in Figure 1. Most lesions in the activity or chronicity indices shared a similar signature within their respective index. In contrast, fibrous crescents displayed an inflammatory signature (CD73, MMP9, MIP1b, and IL-8) despite being part of the NIH chronicity index. Hierarchical clustering based on proteomic signatures revealed that fibrous crescents were more similar to activity-related lesions (Figure 2). Interstitial inflammation (activity) was correlated with biomarkers associated with both active and chronic lesions.
Conclusion: Although fibrous crescents are considered inactive lesions that follow crescentic glomerulonephritis, urine proteomics revealed inflammatory activity associated with fibrous crescents. Several cell types such macrophages, fibroblasts, neutrophils, lymphocytes, and epithelial cells are critical in the formation of crescents. Higher levels of CD73, IL-8 and MMP9 indicate the presence of an inflammatory response involved in glomerular remodeling after extra-capillary proliferation that have an important role in kidney damage. The presence of fibrous crescents in kidney biopsies may indicate ongoing potentially treatable inflammation. Interstitial inflammation, which is linked to worse clinical outcomes, showed a distinct proteomic signature combining both activity and chronicity. A better understanding of the pathophysiology of processes including fibrous crescents and interstitial inflammation is needed to tailor treatment of these pathways leading to chronic damage.
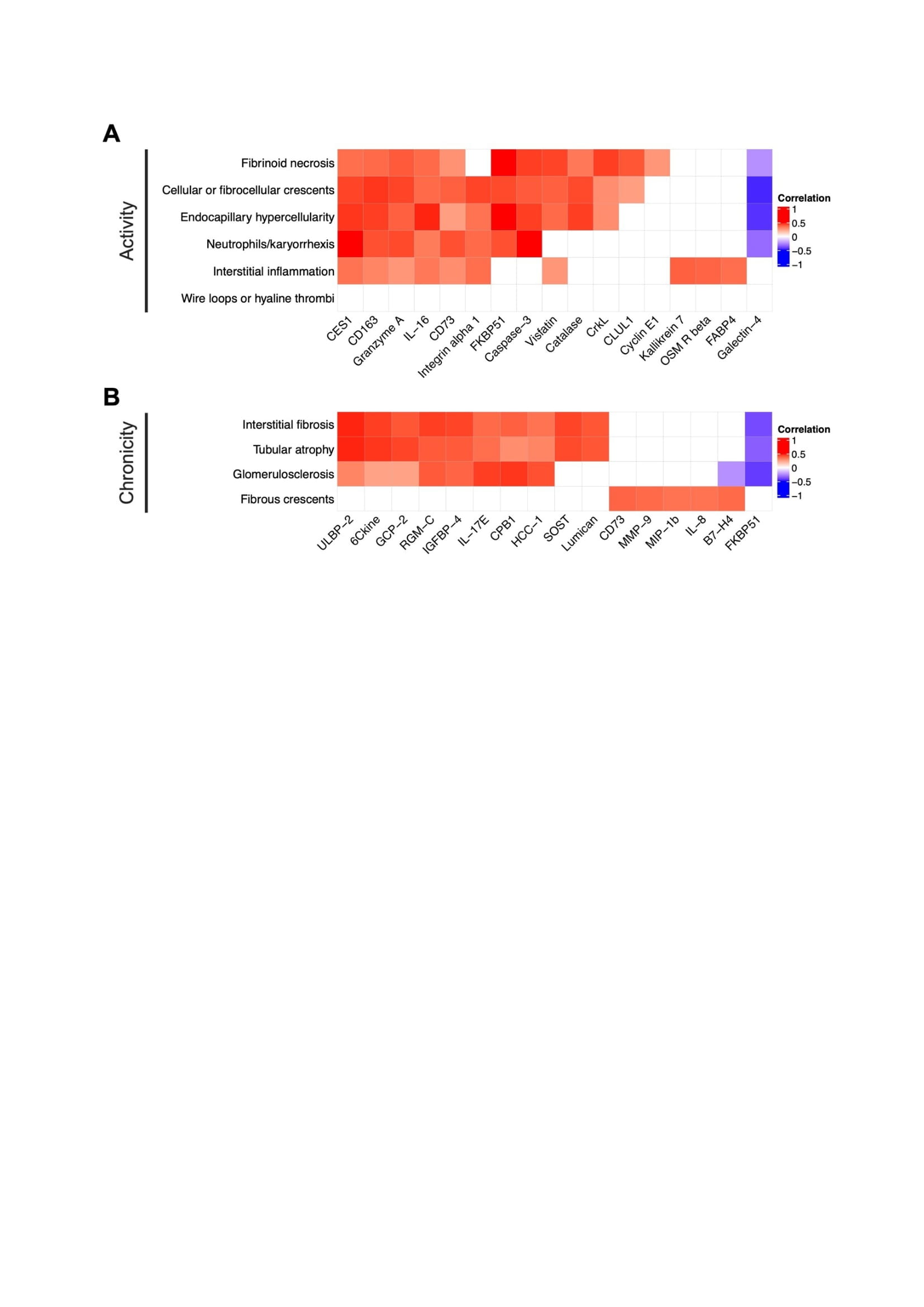 Figure 1. Proteomic signatures of LN histological lesions. Heatmap displaying Spearman correlations of urinary proteins and any single subitem of the NIH activity (A) and chronicity (B) indices. For each lesion, the 5 most correlated proteins are displayed. Empty squares indicate a false discovery (FDR) rate > 0.25.
Figure 1. Proteomic signatures of LN histological lesions. Heatmap displaying Spearman correlations of urinary proteins and any single subitem of the NIH activity (A) and chronicity (B) indices. For each lesion, the 5 most correlated proteins are displayed. Empty squares indicate a false discovery (FDR) rate > 0.25.
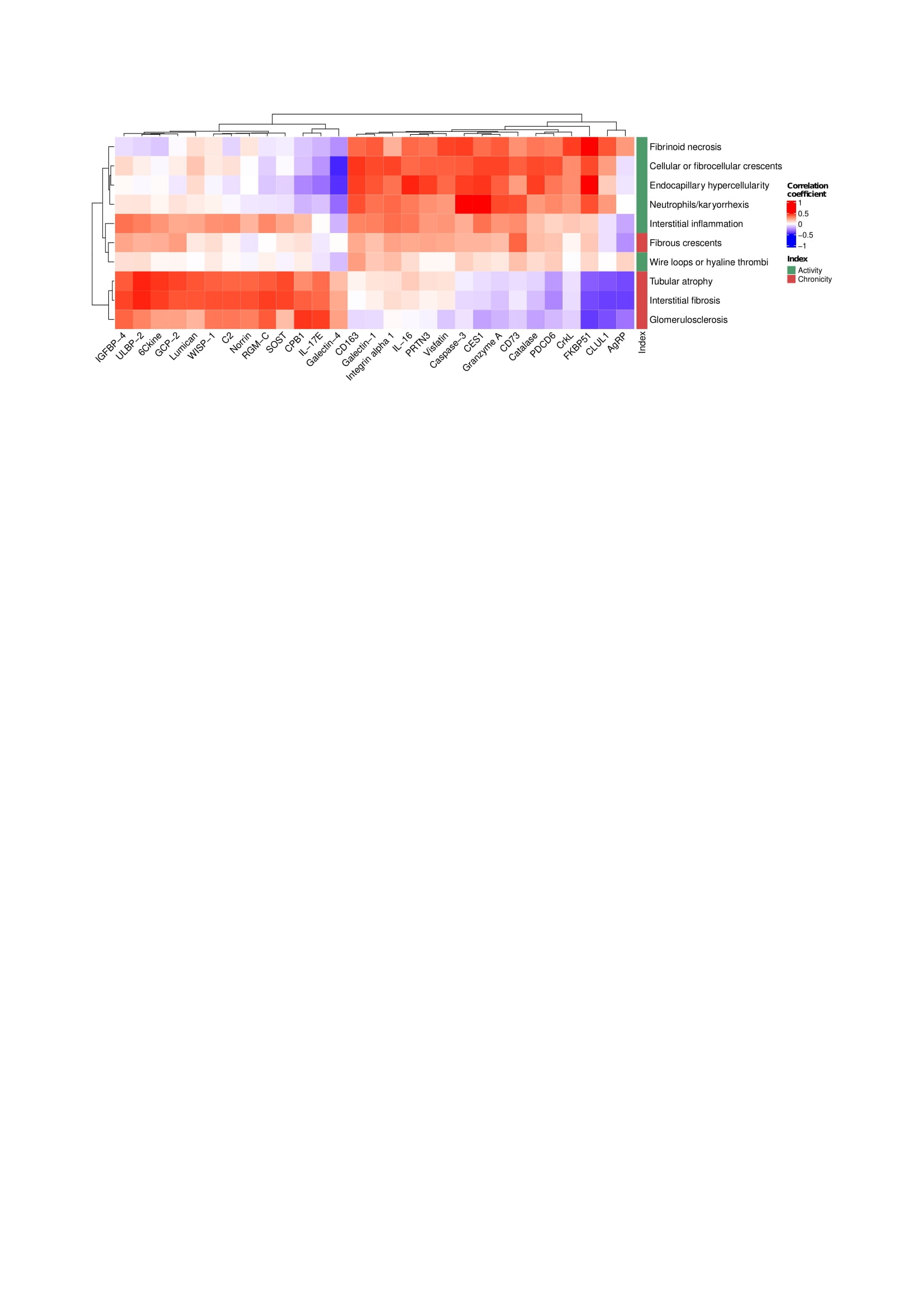 Figure 2. Fibrous crescents cluster with LN activity lesions. Hierarchical clustering based on the correlations of each histological lesion and urinary proteins. All proteins with a strict statistically significant correlation (FDR < 0.01) with at least one histological lesion were included.
Figure 2. Fibrous crescents cluster with LN activity lesions. Hierarchical clustering based on the correlations of each histological lesion and urinary proteins. All proteins with a strict statistically significant correlation (FDR < 0.01) with at least one histological lesion were included.
Disclosures: A. Celia, None; J. Hodgin, None; A. Rosenberg, None; L. Magder, None; J. Buyon, Equillium, GlaxoSmithKlein(GSK), L and M Healthcare Communications, Janssen, Boomcom, Merck/MSD; B. Diamond, None; J. James, Bristol-Myers Squibb(BMS), AstraZeneca, Novartis, Progentec Biosciences; W. Apruzzese, None; P. Fenaroli, None; D. Fine, None; J. Monroy-Trujillo, None; M. Atta, Kira, Horizon, Bristol-Myers Squibb(BMS), REATA, Morphosys AG, Sentien; P. izmirly, None; M. Belmont, None; A. Davidson, None; D. Goldman, None; t. (AMP) RA/SLE, None; M. Petri, Exagen, AstraZeneca, Alexion, Amgen, AnaptysBio, Argenx, Aurinia, Biogen, Caribou Biosciences, CVS Health, EMD Serono, Eli Lilly, Emergent Biosolutions, GlaxoSmithKline (GSK), IQVIA, Janssen, Kira Pharmaceuticals, MedShr, Sanofi, SinoMab, Thermofisher, BPR Scientific Advisory Committee; A. Fava, Sanofi.
Background/Purpose: We employed urine proteomics to define the molecular signatures associated with the histological features quantified by the NIH activity and chronicity indices.
Methods: Glomerular and interstitial lesions in lupus nephritis were quantified (scored 0-3) based on the revised 2018 International Society of Nephrology/Renal Pathology Society (ISN/RPS) classification for lupus nephritis and the modified NIH scoring system by a central renal pathologist (JH). Urinary proteins (1200 biomarkers, RayBiotech Kiloplex) were quantified in urine samples collected on the day of (73%) or within 3 weeks of (27%) the diagnostic kidney biopsy. Proteomic signatures of each lesion were defined based on Spearman correlations of each urine protein with each pathologic lesion.
Results: Ninety-one biopsies were included: 32 (35%) with pure proliferative LN, 33 (36%) with pure membranous LN, and 26 (29%) with mixed LN. The 5 most correlated urinary proteins and each pathologic feature are summarized in Figure 1. Most lesions in the activity or chronicity indices shared a similar signature within their respective index. In contrast, fibrous crescents displayed an inflammatory signature (CD73, MMP9, MIP1b, and IL-8) despite being part of the NIH chronicity index. Hierarchical clustering based on proteomic signatures revealed that fibrous crescents were more similar to activity-related lesions (Figure 2). Interstitial inflammation (activity) was correlated with biomarkers associated with both active and chronic lesions.
Conclusion: Although fibrous crescents are considered inactive lesions that follow crescentic glomerulonephritis, urine proteomics revealed inflammatory activity associated with fibrous crescents. Several cell types such macrophages, fibroblasts, neutrophils, lymphocytes, and epithelial cells are critical in the formation of crescents. Higher levels of CD73, IL-8 and MMP9 indicate the presence of an inflammatory response involved in glomerular remodeling after extra-capillary proliferation that have an important role in kidney damage. The presence of fibrous crescents in kidney biopsies may indicate ongoing potentially treatable inflammation. Interstitial inflammation, which is linked to worse clinical outcomes, showed a distinct proteomic signature combining both activity and chronicity. A better understanding of the pathophysiology of processes including fibrous crescents and interstitial inflammation is needed to tailor treatment of these pathways leading to chronic damage.
 Figure 1. Proteomic signatures of LN histological lesions. Heatmap displaying Spearman correlations of urinary proteins and any single subitem of the NIH activity (A) and chronicity (B) indices. For each lesion, the 5 most correlated proteins are displayed. Empty squares indicate a false discovery (FDR) rate > 0.25.
Figure 1. Proteomic signatures of LN histological lesions. Heatmap displaying Spearman correlations of urinary proteins and any single subitem of the NIH activity (A) and chronicity (B) indices. For each lesion, the 5 most correlated proteins are displayed. Empty squares indicate a false discovery (FDR) rate > 0.25. Figure 2. Fibrous crescents cluster with LN activity lesions. Hierarchical clustering based on the correlations of each histological lesion and urinary proteins. All proteins with a strict statistically significant correlation (FDR < 0.01) with at least one histological lesion were included.
Figure 2. Fibrous crescents cluster with LN activity lesions. Hierarchical clustering based on the correlations of each histological lesion and urinary proteins. All proteins with a strict statistically significant correlation (FDR < 0.01) with at least one histological lesion were included.Disclosures: A. Celia, None; J. Hodgin, None; A. Rosenberg, None; L. Magder, None; J. Buyon, Equillium, GlaxoSmithKlein(GSK), L and M Healthcare Communications, Janssen, Boomcom, Merck/MSD; B. Diamond, None; J. James, Bristol-Myers Squibb(BMS), AstraZeneca, Novartis, Progentec Biosciences; W. Apruzzese, None; P. Fenaroli, None; D. Fine, None; J. Monroy-Trujillo, None; M. Atta, Kira, Horizon, Bristol-Myers Squibb(BMS), REATA, Morphosys AG, Sentien; P. izmirly, None; M. Belmont, None; A. Davidson, None; D. Goldman, None; t. (AMP) RA/SLE, None; M. Petri, Exagen, AstraZeneca, Alexion, Amgen, AnaptysBio, Argenx, Aurinia, Biogen, Caribou Biosciences, CVS Health, EMD Serono, Eli Lilly, Emergent Biosolutions, GlaxoSmithKline (GSK), IQVIA, Janssen, Kira Pharmaceuticals, MedShr, Sanofi, SinoMab, Thermofisher, BPR Scientific Advisory Committee; A. Fava, Sanofi.

