Back
Abstract Session
Session: Abstracts: RA – Diagnosis, Manifestations, and Outcomes III: RA ILD (2248–2253)
2253: Sputum Cytokines Are Elevated in Rheumatoid Arthritis-Associated Interstitial Lung Disease
Monday, November 14, 2022
5:45 PM – 5:55 PM Eastern Time
Location: Room 103
- TW
Timothy Wilson, MD
Thomas Jefferson University
Philadelphia, PA, United States
Presenting Author(s)
Timothy Wilson1, Kevin D Deane2, Jonathan Dekermanjian2, Joyce Lee2, Marie Feser3, Stephen Humphries4, Joshua Solomon4 and Kristen Demoruelle3, 1University of Colorado Denver Anschutz Medical Campus, Aurora, CO, 2University of Colorado Denver Anschutz Medical Campus, Denver, CO, 3University of Colorado Anschutz Medical Campus, Aurora, CO, 4National Jewish Health, Denver, CO
Background/Purpose: Interstitial lung disease (ILD) is a well-recognized comorbidity in rheumatoid arthritis (RA) that contributes significantly to morbidity and mortality. ILD is diagnosed in up to 10% of RA patients but up to 40% of patients have subclinical lung parenchymal abnormalities without a clinical diagnosis of ILD. The mechanisms driving the development of RA-ILD including the progression of subclinical to established ILD remain poorly understood. Identifying sputum-based biomarkers, which are likely more reflective of the pathobiology in RA-ILD compared to blood-based biomarkers, may be more informative in characterizing the disease pathogenesis.
Methods: We evaluated 34 RA patients without ILD confirmed by high-resolution CT (HRCT) (RA-no-ILD), 18 RA patients with a clinical diagnosis of ILD as reviewed by a pulmonologist specializing in ILD (RA-ILD), and 24 RA patients with subclinical ILD defined as lung parenchymal abnormalities on HRCT but normal pulmonary physiology (RA-sub-ILD). Induced sputum was collected from all patients and HRCT and pulmonary function tests were completed within 3 months of sputum collection. Lung fibrosis was quantified by HRCT using data-driven textural analysis (DTA) fibrosis scores. Sputum supernatant was tested for levels of 31 cytokines/chemokines using multiplex assays (Meso Scale Discovery). A Random Forest (RForest) classifier model using sputum cytokines/chemokines was used to discriminate between RA-no-ILD and RA-ILD. DNA was extracted from participants with available serum to genotype for a single nucleotide polymorphism (rs35705950) within the promoter region of the MUC5B gene (Thermo Fischer) as the minor "T" allele has been shown to be one of the strongest risk factors for developing ILD in RA.
Results: Within the RForest model, transforming growth factor-beta 1 (TGF-β1), tumor necrosis factor-alpha (TNF-α), monocyte chemoattractant protein-1 (MCP-1), and interleukin-4 (IL-4) were the most impactful in classifying a patient as RA-ILD vs. RA-no-ILD (Figure 1); furthermore, these cytokines were significantly elevated in RA-ILD using multivariate regression models (Figure 2). TGF-β1 and TNF-α were also significantly elevated in RA-sub-ILD compared to RA-no-ILD (p=0.03 and p=0.05, respectively, Figure 2). TGF-β1 levels in RA-sub-ILD subjects were positively correlated with DTA fibrosis score. TNF-α levels in RA-ILD subjects were positively correlated with diffusion capacity of carbon monoxide (DLCO). Lastly, within RA-ILD, there was a trend toward higher cytokine levels in those with the at-risk MUC5B "T" allele compared to those homozygous for the wild type "G" allele (Figure 3).
Conclusion: We demonstrate that sputum levels of TGF-β1, TNF-α, MCP-1, and IL-4 are significantly elevated in RA-ILD and a portion of these cytokines are also increased in subclinical disease and correlated with lung physiology. In pilot testing, we also found a trend of higher sputum cytokine levels in RA-ILD subjects with the MUC5B promoter variant. Future studies are needed to better understand the role of these cytokines in ILD and specifically evaluate the influence of MUC5B genetics on cytokine generation in the lung and the development of RA-ILD.
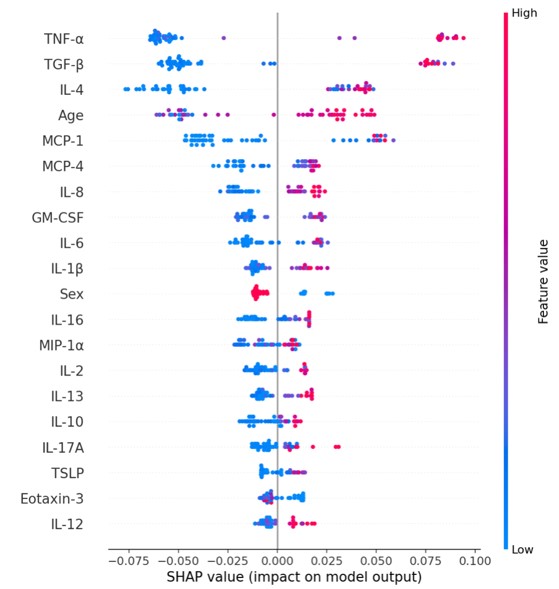 Figure 1. Sputum cytokines in RA-ILD. Top 20 cytokines/chemokines ranked from most (top) to least (bottom) impactful in predicting whether a subject is RA-no-ILD vs RA-ILD . Each data point represents one subject. Shapley Additive Explanation (SHAP) value depicts the probability of the impact on the model’s prediction. The feature value depicts whether a variable has a low, medium or high value.
Figure 1. Sputum cytokines in RA-ILD. Top 20 cytokines/chemokines ranked from most (top) to least (bottom) impactful in predicting whether a subject is RA-no-ILD vs RA-ILD . Each data point represents one subject. Shapley Additive Explanation (SHAP) value depicts the probability of the impact on the model’s prediction. The feature value depicts whether a variable has a low, medium or high value.
<img src=https://www.abstractscorecard.com/uploads/Tasks/upload/17574/QHOPTGBB-1271487-2-ANY.jpg width=440 height=416.634482758621 border=0 style=border-style: none;>
.jpg) Figure 3. Correlation of cytokine levels with MUC5B promoter variant. MUC5B genotype (“G” - wild type allele, “T” – minor, at-risk allele) correlated with sputum levels of TGF-β1 (panel A), IL-4 (panel B), MCP-1 (panel C), and TNF-α (panel D) in RA-ILD subjects (n=16). P-value based on nonparametric Mann-Whitney U testing.
Figure 3. Correlation of cytokine levels with MUC5B promoter variant. MUC5B genotype (“G” - wild type allele, “T” – minor, at-risk allele) correlated with sputum levels of TGF-β1 (panel A), IL-4 (panel B), MCP-1 (panel C), and TNF-α (panel D) in RA-ILD subjects (n=16). P-value based on nonparametric Mann-Whitney U testing.
Disclosures: T. Wilson, None; K. Deane, Werfen; J. Dekermanjian, None; J. Lee, None; M. Feser, None; S. Humphries, None; J. Solomon, None; K. Demoruelle, Boehringer-Ingelheim, Pfizer.
Background/Purpose: Interstitial lung disease (ILD) is a well-recognized comorbidity in rheumatoid arthritis (RA) that contributes significantly to morbidity and mortality. ILD is diagnosed in up to 10% of RA patients but up to 40% of patients have subclinical lung parenchymal abnormalities without a clinical diagnosis of ILD. The mechanisms driving the development of RA-ILD including the progression of subclinical to established ILD remain poorly understood. Identifying sputum-based biomarkers, which are likely more reflective of the pathobiology in RA-ILD compared to blood-based biomarkers, may be more informative in characterizing the disease pathogenesis.
Methods: We evaluated 34 RA patients without ILD confirmed by high-resolution CT (HRCT) (RA-no-ILD), 18 RA patients with a clinical diagnosis of ILD as reviewed by a pulmonologist specializing in ILD (RA-ILD), and 24 RA patients with subclinical ILD defined as lung parenchymal abnormalities on HRCT but normal pulmonary physiology (RA-sub-ILD). Induced sputum was collected from all patients and HRCT and pulmonary function tests were completed within 3 months of sputum collection. Lung fibrosis was quantified by HRCT using data-driven textural analysis (DTA) fibrosis scores. Sputum supernatant was tested for levels of 31 cytokines/chemokines using multiplex assays (Meso Scale Discovery). A Random Forest (RForest) classifier model using sputum cytokines/chemokines was used to discriminate between RA-no-ILD and RA-ILD. DNA was extracted from participants with available serum to genotype for a single nucleotide polymorphism (rs35705950) within the promoter region of the MUC5B gene (Thermo Fischer) as the minor "T" allele has been shown to be one of the strongest risk factors for developing ILD in RA.
Results: Within the RForest model, transforming growth factor-beta 1 (TGF-β1), tumor necrosis factor-alpha (TNF-α), monocyte chemoattractant protein-1 (MCP-1), and interleukin-4 (IL-4) were the most impactful in classifying a patient as RA-ILD vs. RA-no-ILD (Figure 1); furthermore, these cytokines were significantly elevated in RA-ILD using multivariate regression models (Figure 2). TGF-β1 and TNF-α were also significantly elevated in RA-sub-ILD compared to RA-no-ILD (p=0.03 and p=0.05, respectively, Figure 2). TGF-β1 levels in RA-sub-ILD subjects were positively correlated with DTA fibrosis score. TNF-α levels in RA-ILD subjects were positively correlated with diffusion capacity of carbon monoxide (DLCO). Lastly, within RA-ILD, there was a trend toward higher cytokine levels in those with the at-risk MUC5B "T" allele compared to those homozygous for the wild type "G" allele (Figure 3).
Conclusion: We demonstrate that sputum levels of TGF-β1, TNF-α, MCP-1, and IL-4 are significantly elevated in RA-ILD and a portion of these cytokines are also increased in subclinical disease and correlated with lung physiology. In pilot testing, we also found a trend of higher sputum cytokine levels in RA-ILD subjects with the MUC5B promoter variant. Future studies are needed to better understand the role of these cytokines in ILD and specifically evaluate the influence of MUC5B genetics on cytokine generation in the lung and the development of RA-ILD.
 Figure 1. Sputum cytokines in RA-ILD. Top 20 cytokines/chemokines ranked from most (top) to least (bottom) impactful in predicting whether a subject is RA-no-ILD vs RA-ILD . Each data point represents one subject. Shapley Additive Explanation (SHAP) value depicts the probability of the impact on the model’s prediction. The feature value depicts whether a variable has a low, medium or high value.
Figure 1. Sputum cytokines in RA-ILD. Top 20 cytokines/chemokines ranked from most (top) to least (bottom) impactful in predicting whether a subject is RA-no-ILD vs RA-ILD . Each data point represents one subject. Shapley Additive Explanation (SHAP) value depicts the probability of the impact on the model’s prediction. The feature value depicts whether a variable has a low, medium or high value. <img src=https://www.abstractscorecard.com/uploads/Tasks/upload/17574/QHOPTGBB-1271487-2-ANY.jpg width=440 height=416.634482758621 border=0 style=border-style: none;>
Figure 2. Top 4 cytokine levels in RA-sub-ILD and RA-ILD. Sputum levels in all groups of top 4 most influential cytokines as identified in RForest model (TGF-β1, panel A; TNF-α, panel B; MCP-1, panel C; and IL-4, panel D). ** = p < 0.05, ns = p>0.05; p-value based on multivariate regression model adjusting for co-variates (age, sex, smoking status).
.jpg) Figure 3. Correlation of cytokine levels with MUC5B promoter variant. MUC5B genotype (“G” - wild type allele, “T” – minor, at-risk allele) correlated with sputum levels of TGF-β1 (panel A), IL-4 (panel B), MCP-1 (panel C), and TNF-α (panel D) in RA-ILD subjects (n=16). P-value based on nonparametric Mann-Whitney U testing.
Figure 3. Correlation of cytokine levels with MUC5B promoter variant. MUC5B genotype (“G” - wild type allele, “T” – minor, at-risk allele) correlated with sputum levels of TGF-β1 (panel A), IL-4 (panel B), MCP-1 (panel C), and TNF-α (panel D) in RA-ILD subjects (n=16). P-value based on nonparametric Mann-Whitney U testing.Disclosures: T. Wilson, None; K. Deane, Werfen; J. Dekermanjian, None; J. Lee, None; M. Feser, None; S. Humphries, None; J. Solomon, None; K. Demoruelle, Boehringer-Ingelheim, Pfizer.

