Back
Poster Session B
Systemic lupus erythematosus (SLE)
Session: (0629–0670) SLE – Etiology and Pathogenesis Poster
0660: Serum Proteins and Metabolites Differ in SLE Patients with Coronary Microvascular Dysfunction (CMD) as Determined by Cardiac MRI Compared to SLE Patients Without CMD – a Pilot Study
Sunday, November 13, 2022
9:00 AM – 10:30 AM Eastern Time
Location: Virtual Poster Hall
- AH
Audrey Hagiwara, MD
Cedars-Sinai Medical Center
Los Angeles, CA, United States
Abstract Poster Presenter(s)
Audrey Hagiwara1, Moumita Bose1, Aleksandr Stotland1, Aleksandra Binek1, Koen Raedschelders1, Janet Wei1, C. Noel Bairey Merz1, Sarah Parker1, Jennifer Van Eyk1, Daniel Wallace1, Mariko Ishimori2 and Caroline Jefferies1, 1Cedars-Sinai Medical Center, Los Angeles, CA, 2Cedars-Sinai Health System, Los Angeles, CA
Background/Purpose: Cardiovascular disease (CVD) is a significant cause of mortality in patients with SLE. While obstructive coronary artery disease (CAD) is a leading cause of cardiac death in SLE, newer evidence suggests that other cardiac manifestations may contribute significantly to mortality, including coronary microvascular dysfunction (CMD) – a form of ischemia without obstructive CAD (Weber et al., 2021). The presence of CMD in SLE is associated with decreased total anti-oxidant levels and increased CRP levels suggesting a role for oxidative stress and inflammation in the pathogenesis of CMD in lupus (Yilmaz et al., 2012). We performed a comprehensive proteomic analysis of serum protein and metabolite profiles in SLE participants with and without CMD on cardiac MRI (cMRI).
Methods: SLE patients (n=11, all met ACR 1997 criteria, Table) recruited to the study underwent cMRI using our published protocol (Ishimori et al., 2011). Metabolites were extracted from patient serum samples and analyzed by LC-MS/MS using a targeted dynamic multiple reaction monitoring (dMRM) method in combination with the MassHunter Metabolomics pipeline to profile the 218 metabolites of the central carbon chain (Agilent Technologies). Serum proteins were analyzed by data-independent acquisition (DIA) LC-MS/MS and analyzed using mapDIA software. Statistical analysis, functional pathway characterization and visualization for both assays were performed as described elsewhere (Mc Ardle et al., 2022; Stotland et al., 2020).
Results: Overall, 5/11 participants had myocardial perfusion reserve index (MPRI) values < 1.84, consistent with CMD. We identified 19 metabolites differentially abundant in SLE patients with (p < 0.05) compared to SLE participants without CMD (Figure 1), including 18 that were significantly upregulated in CMD patients, and 1 (2-deoxyguanosine 5-diphosphate) was down-regulated. Pathway analysis performed using REACTOME identified a number of metabolic and biosynthetic pathways affected between the two groups, including pyrimidine biosynthesis (p = 2.9e-04, FDR 0.037), vitamin B2 metabolism (p = 0.005, FDR 0.037), and creatine metabolism (p = 0.004, FDR 0.037). Interestingly we found 10 serum proteins negatively correlated with MPRI on cMRI (Figure 2) and 4 positively. Of those that negatively correlated, epidermal growth factor receptor (EGFR, r = -0.661, p = 0.027) and leucine rich alpha-2 glycoprotein 1 (A2GL, r = -0.675, p = 0.023) were the most noteworthy, as both have known associations with increased CVD risk. Clusterin (CLUS) was the most negatively correlated of all the proteins with MPRI (r = -0.922, p = 5.55e-005), which is interesting given the strong association of clusterin with elevated CRP levels and other markers of cardiovascular risk (Wittwer & Bradley, 2021).
Conclusion: SLE participants with CMD demonstrated significant upregulation of metabolites, which suggests alterations in metabolic pathways and substrate utilization may contribute to the CMD pathophysiology in SLE. Additionally, these proteomic data support increased expression of biomarkers previously shown to be associated with CVD. These results may help elucidate the mechanisms driving the development of CMD in SLE.
.jpg) Table: Demographic and Characteristics of SLE Participants
Table: Demographic and Characteristics of SLE Participants
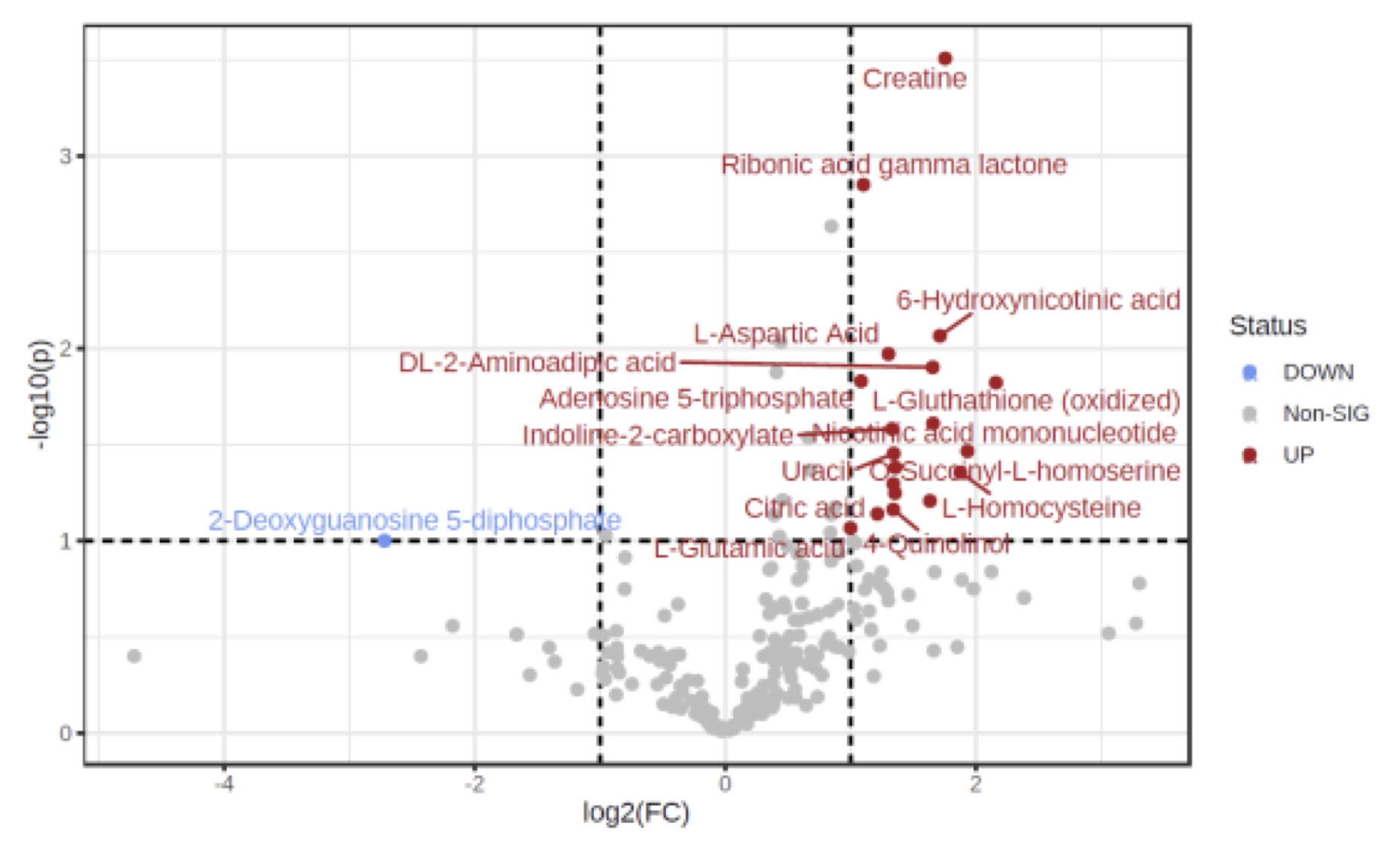 Figure 1: Important features selected by volcano plot with fold change threshold (x) 2 and t-tests
Figure 1: Important features selected by volcano plot with fold change threshold (x) 2 and t-tests
threshold (y) 0.1. The red circles represent features above the threshold.
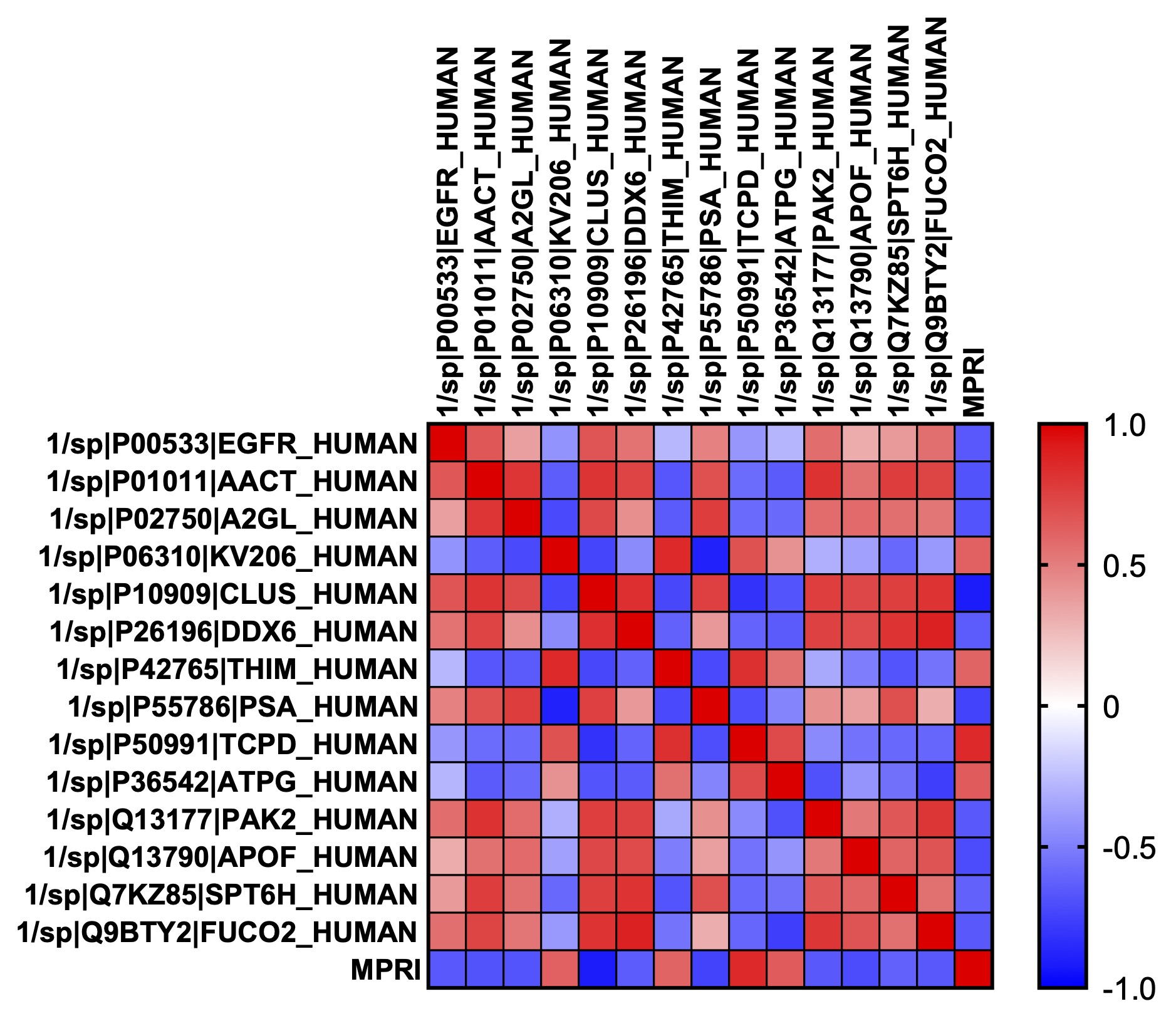 Figure 2: Heatmap of Pearson’s correlation coefficient between each serum protein found to be significantly correlated with MPRI. The correlation between each of the proteins with MPRI is down on the bottom row. In each case, p < 0.05. Epidermal growth factor receptor (EGFR); Alpha-1-antichymotrypsin (AACT); Leucine rich alpha-2 glycoprotein 1 (A2GL); Ig kappa chain V-II region (KV206); Clusterin (CLUS); ATP-dependent RNA helicase (DDX6); Mitochondrial 3-ketoacyl-CoA thiolase (THIM); Puromycin-sensitive aminopeptidase (PSA); T-complex protein 1 subunit delta (TCPD); Mitochondrial ATP synthase subunit gamma (ATPG); Serine/threonine-protein kinase (PAK2); Apolipoprotein F (APOF); Transcription elongation factor SPT6 (SPT6H); Plasma alpha-L-fucosidase (FUCO2).
Figure 2: Heatmap of Pearson’s correlation coefficient between each serum protein found to be significantly correlated with MPRI. The correlation between each of the proteins with MPRI is down on the bottom row. In each case, p < 0.05. Epidermal growth factor receptor (EGFR); Alpha-1-antichymotrypsin (AACT); Leucine rich alpha-2 glycoprotein 1 (A2GL); Ig kappa chain V-II region (KV206); Clusterin (CLUS); ATP-dependent RNA helicase (DDX6); Mitochondrial 3-ketoacyl-CoA thiolase (THIM); Puromycin-sensitive aminopeptidase (PSA); T-complex protein 1 subunit delta (TCPD); Mitochondrial ATP synthase subunit gamma (ATPG); Serine/threonine-protein kinase (PAK2); Apolipoprotein F (APOF); Transcription elongation factor SPT6 (SPT6H); Plasma alpha-L-fucosidase (FUCO2).
Disclosures: A. Hagiwara, None; M. Bose, None; A. Stotland, None; A. Binek, None; K. Raedschelders, None; J. Wei, None; C. Bairey Merz, None; S. Parker, None; J. Van Eyk, None; D. Wallace, None; M. Ishimori, None; C. Jefferies, None.
Background/Purpose: Cardiovascular disease (CVD) is a significant cause of mortality in patients with SLE. While obstructive coronary artery disease (CAD) is a leading cause of cardiac death in SLE, newer evidence suggests that other cardiac manifestations may contribute significantly to mortality, including coronary microvascular dysfunction (CMD) – a form of ischemia without obstructive CAD (Weber et al., 2021). The presence of CMD in SLE is associated with decreased total anti-oxidant levels and increased CRP levels suggesting a role for oxidative stress and inflammation in the pathogenesis of CMD in lupus (Yilmaz et al., 2012). We performed a comprehensive proteomic analysis of serum protein and metabolite profiles in SLE participants with and without CMD on cardiac MRI (cMRI).
Methods: SLE patients (n=11, all met ACR 1997 criteria, Table) recruited to the study underwent cMRI using our published protocol (Ishimori et al., 2011). Metabolites were extracted from patient serum samples and analyzed by LC-MS/MS using a targeted dynamic multiple reaction monitoring (dMRM) method in combination with the MassHunter Metabolomics pipeline to profile the 218 metabolites of the central carbon chain (Agilent Technologies). Serum proteins were analyzed by data-independent acquisition (DIA) LC-MS/MS and analyzed using mapDIA software. Statistical analysis, functional pathway characterization and visualization for both assays were performed as described elsewhere (Mc Ardle et al., 2022; Stotland et al., 2020).
Results: Overall, 5/11 participants had myocardial perfusion reserve index (MPRI) values < 1.84, consistent with CMD. We identified 19 metabolites differentially abundant in SLE patients with (p < 0.05) compared to SLE participants without CMD (Figure 1), including 18 that were significantly upregulated in CMD patients, and 1 (2-deoxyguanosine 5-diphosphate) was down-regulated. Pathway analysis performed using REACTOME identified a number of metabolic and biosynthetic pathways affected between the two groups, including pyrimidine biosynthesis (p = 2.9e-04, FDR 0.037), vitamin B2 metabolism (p = 0.005, FDR 0.037), and creatine metabolism (p = 0.004, FDR 0.037). Interestingly we found 10 serum proteins negatively correlated with MPRI on cMRI (Figure 2) and 4 positively. Of those that negatively correlated, epidermal growth factor receptor (EGFR, r = -0.661, p = 0.027) and leucine rich alpha-2 glycoprotein 1 (A2GL, r = -0.675, p = 0.023) were the most noteworthy, as both have known associations with increased CVD risk. Clusterin (CLUS) was the most negatively correlated of all the proteins with MPRI (r = -0.922, p = 5.55e-005), which is interesting given the strong association of clusterin with elevated CRP levels and other markers of cardiovascular risk (Wittwer & Bradley, 2021).
Conclusion: SLE participants with CMD demonstrated significant upregulation of metabolites, which suggests alterations in metabolic pathways and substrate utilization may contribute to the CMD pathophysiology in SLE. Additionally, these proteomic data support increased expression of biomarkers previously shown to be associated with CVD. These results may help elucidate the mechanisms driving the development of CMD in SLE.
.jpg) Table: Demographic and Characteristics of SLE Participants
Table: Demographic and Characteristics of SLE Participants Figure 1: Important features selected by volcano plot with fold change threshold (x) 2 and t-tests
Figure 1: Important features selected by volcano plot with fold change threshold (x) 2 and t-teststhreshold (y) 0.1. The red circles represent features above the threshold.
 Figure 2: Heatmap of Pearson’s correlation coefficient between each serum protein found to be significantly correlated with MPRI. The correlation between each of the proteins with MPRI is down on the bottom row. In each case, p < 0.05. Epidermal growth factor receptor (EGFR); Alpha-1-antichymotrypsin (AACT); Leucine rich alpha-2 glycoprotein 1 (A2GL); Ig kappa chain V-II region (KV206); Clusterin (CLUS); ATP-dependent RNA helicase (DDX6); Mitochondrial 3-ketoacyl-CoA thiolase (THIM); Puromycin-sensitive aminopeptidase (PSA); T-complex protein 1 subunit delta (TCPD); Mitochondrial ATP synthase subunit gamma (ATPG); Serine/threonine-protein kinase (PAK2); Apolipoprotein F (APOF); Transcription elongation factor SPT6 (SPT6H); Plasma alpha-L-fucosidase (FUCO2).
Figure 2: Heatmap of Pearson’s correlation coefficient between each serum protein found to be significantly correlated with MPRI. The correlation between each of the proteins with MPRI is down on the bottom row. In each case, p < 0.05. Epidermal growth factor receptor (EGFR); Alpha-1-antichymotrypsin (AACT); Leucine rich alpha-2 glycoprotein 1 (A2GL); Ig kappa chain V-II region (KV206); Clusterin (CLUS); ATP-dependent RNA helicase (DDX6); Mitochondrial 3-ketoacyl-CoA thiolase (THIM); Puromycin-sensitive aminopeptidase (PSA); T-complex protein 1 subunit delta (TCPD); Mitochondrial ATP synthase subunit gamma (ATPG); Serine/threonine-protein kinase (PAK2); Apolipoprotein F (APOF); Transcription elongation factor SPT6 (SPT6H); Plasma alpha-L-fucosidase (FUCO2).Disclosures: A. Hagiwara, None; M. Bose, None; A. Stotland, None; A. Binek, None; K. Raedschelders, None; J. Wei, None; C. Bairey Merz, None; S. Parker, None; J. Van Eyk, None; D. Wallace, None; M. Ishimori, None; C. Jefferies, None.

