Back
Abstract Session
Fibrosing rheumatic diseases (scleroderma, MCTD, IgG4-related disease, scleroderma mimics)
Session: Abstracts: Systemic Sclerosis and Related Disorders – Basic Science (1621–1624)
1622: TNF Receptor 1 Drives Murine Pulmonary Arterial Hypertension and Is Characterized by Loss of Capillary Endothelial Cells and Pericytes, Smooth Muscle Cell Proliferation, and Alterations in Fibroblast Phenotype
Sunday, November 13, 2022
5:15 PM – 5:25 PM Eastern Time
Location: Room 201
.png)
Benjamin D. Korman, MD
University of Rochester
Rochester, NY, United States
Presenting Author(s)
Stacey Duemmel1, Marc Nuzzo1, Soumyaroop Bhattacharya1, Qingfu Xu1, Amy Mohan1 and Benjamin Korman2, 1URMC, Rochester, NY, 2University of Rochester, Rochester, NY
Background/Purpose: We previously demonstrated that TNF-transgenic (TNF-Tg) mice have findings consistent with connective-tissue disease associated pulmonary arterial hypertension (CTD-PAH), and that this pathology is mediated by non-hematopoietic cells. Because other TNF-transgenic models have shown divergent TNF receptor pathology between arthritis and visceral disease, we sought to understand the contribution of TNF receptors 1 and 2 (TNFR1 and TNFR2) in PAH pathogenesis. Moreover, to better understand the cellular mechanism contributing to PAH in this model, we sought to evaluate the roles of endothelial and stromal cell alterations in lungs from TNF-Tg mice.
Methods: TNF-Tg mice were crossed with mice deficient in either TNFR1 or TNFR2. Single and double-transgenic mice (5-6 per group) were assessed at 4.5 months of age for pulmonary vascular alterations by histomorphometry and micro-CT angiography, right ventricular pathology by histology, echocardiography, and right heart catheterization. Joint erosion was assessed clinically and by micro-CT. Lungs for WT and TNF-Tg mice (n=3 per timepoint per group) were harvested at 1.5, 3.5, and 5 months of age and were digested to a single cell suspension. Flow cytometry was used to deplete hematopoietic (CD45+) and epithelial (CD326+) cells and single cell RNA sequencing (scRNAseq) was performed on the resulting endothelial and mesenchymal cells. Library preparation was done on enzymatically digested cells (n=45,4335) using a 10X Genomics® Chromium instrument and sequencing was performed on an Illumina NovaSeq6000. ScRNA-seq reads were examined for quality and transcripts were mapped to reference mouse genome and assigned to cells of origin using the TopCell atlas. Analysis was performed using Seurat v3.4 to identify, cluster, and visualize distinct cell populations by PCA and UMAP methodologies and to perform differential gene expression analysis.
Results: TNFR1-KO/TNF-Tg mice were protected from PAH and arthritis by clinical, histologic, imaging, and hemodynamic assessment whereas TNFR2-KO/TNF-Tg mice had disease which was indistinguishable from TNF-Tg mice (Figure 1). Single cell RNA-sequencing of lungs revealed that TNF-Tg mice had reduced endothelial cell numbers, particularly gCAP cells, and that TNF-Tg endothelial cells lost canonical endothelial markers (vWF) but over-expressed genes associated with precursors (CD34), inflammation (CD74), adhesion (VCAM1), and the basement membrane (HSPG2, COL4A1). (Figure 2) Additionally, analysis of stromal cells demonstrated that TNF-Tg mice accumulated smooth muscle cells with a proliferative/invasive phenotype, lost pericytes, and showed shifts in fibroblasts with loss of lipofibrobasts and myofibroblasts and increases in matrix fibroblasts. (Figure 3).
Conclusion: TNFR1 rather than TNFR2 is responsible for TNF mediated pulmonary endothelial and stromal pathology which results in PAH in a mouse model of CTD-PAH. Single cell RNA sequencing revealed important TNF-driven changes in cellular phenotype in the endothelial, smooth muscle, pericyte, and fibroblast lineages.
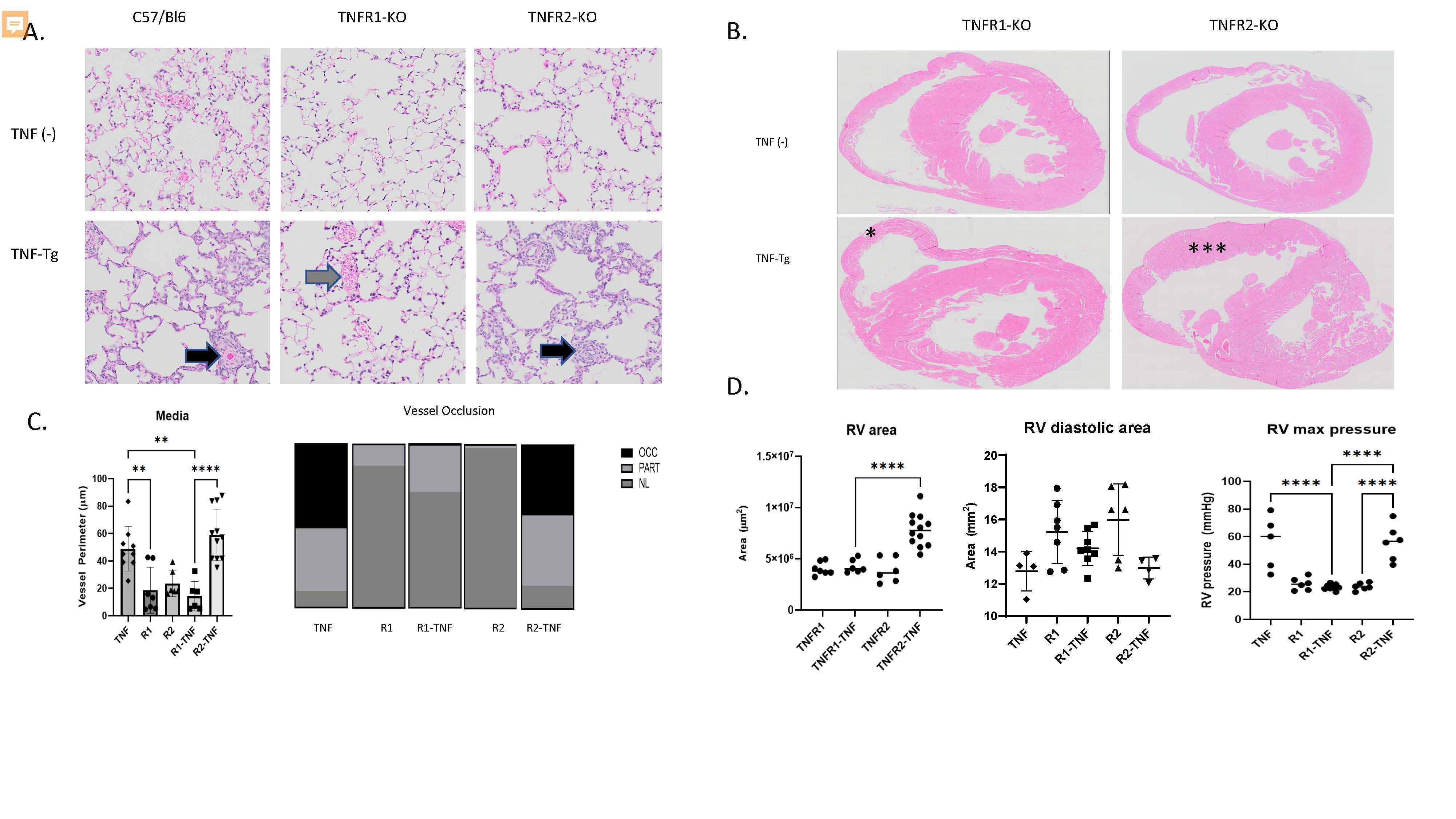 Figure 1. TNFR1 mediates PAH and arthritis phenotype in a mouse model of CTD-PAH. A. Lung histology (H&E, 20x) images demonstrate that TNF-Tg and TNFR2-KO/TNF-Tg mice, but not TNFR1-KO/TNF-Tg mice have pulmonary inflammation and vascular occlusion (arrowheads). B. Cardiac histology (H&E, 4x) demonstrates RV hypertrophy in TNFR2-KO/TNF-Tg (**) mice but not TNFR1-KO/TNF-Tg mice (*). C. (Left) Quantification of pulmonary vessel media perimeter (Right) Proportion of partially and fully occluded vessels in each phenotype. D. (Left) RV area measured by histomorphometry, (Middle) RV end diastolic area measured by echocardiography (Right) RV systolic pressure measured by right heart catheterization.
Figure 1. TNFR1 mediates PAH and arthritis phenotype in a mouse model of CTD-PAH. A. Lung histology (H&E, 20x) images demonstrate that TNF-Tg and TNFR2-KO/TNF-Tg mice, but not TNFR1-KO/TNF-Tg mice have pulmonary inflammation and vascular occlusion (arrowheads). B. Cardiac histology (H&E, 4x) demonstrates RV hypertrophy in TNFR2-KO/TNF-Tg (**) mice but not TNFR1-KO/TNF-Tg mice (*). C. (Left) Quantification of pulmonary vessel media perimeter (Right) Proportion of partially and fully occluded vessels in each phenotype. D. (Left) RV area measured by histomorphometry, (Middle) RV end diastolic area measured by echocardiography (Right) RV systolic pressure measured by right heart catheterization.
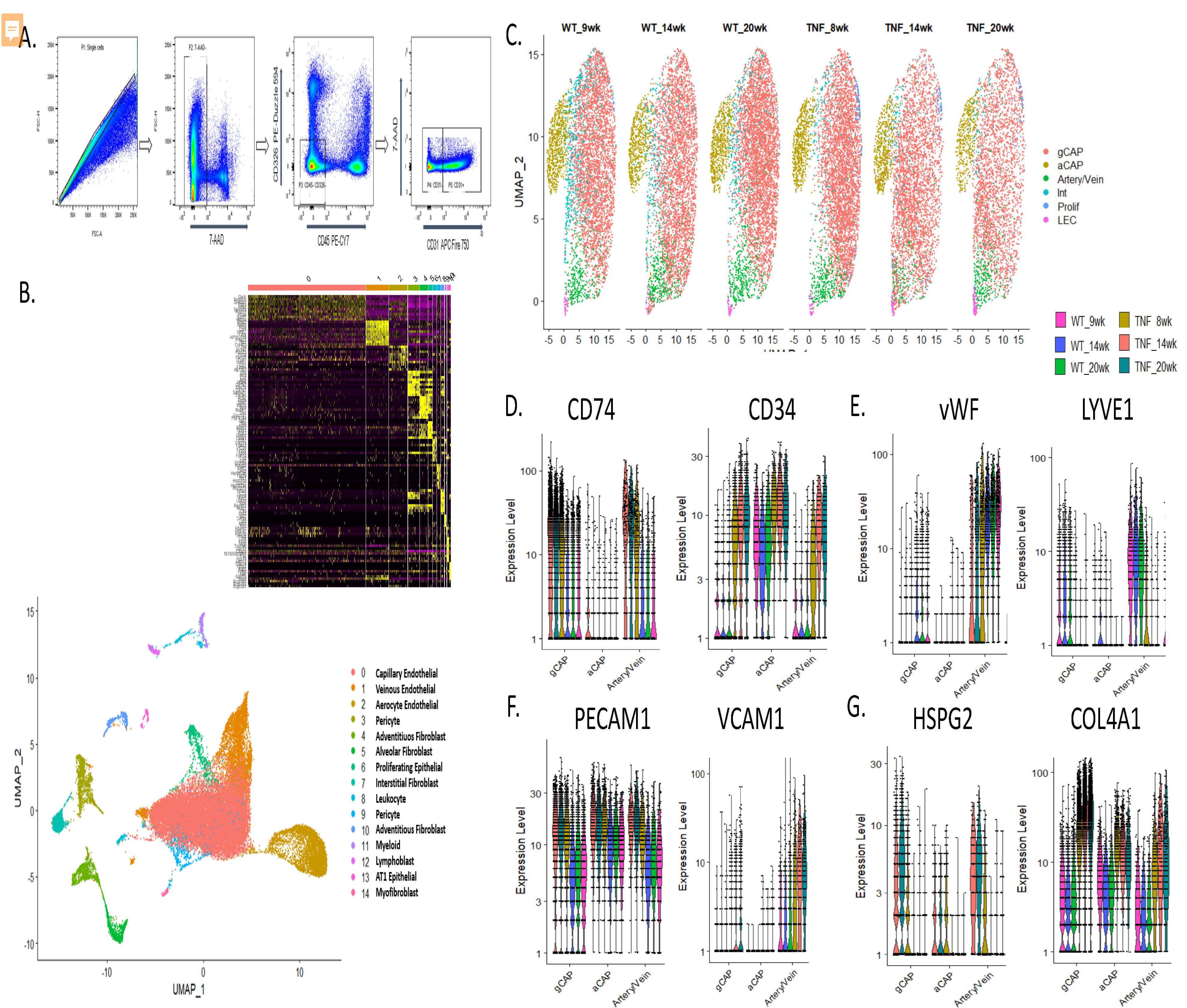 Figure 2. Single cell digestion, sorting, and characterization of endothelial cell phenotypes in TNF-Tg lungs. A. Flow cytometric gating strategy to deplete lungs of CD45+ and CD326+ cells. B. uMAP projection demonstrating 15 pulmonary endothelial and menenchymal cellular populations identified after single cell RNA sequencing across 43,455 cells sequenced. (inset upper) heatmap demonstrating canonical genes used to identify cellular sub-populations C. uMAP projection demonstrating subsets of endothelial cells, note decreased cellular density of general capillary endothelial cells (gCAP) particularly at the 14 and 20 week timepoints in TNF-Tg mice. D. Up-regulation of CD34 and down-regulation of CD74, markers previously shown to be differentially regulated in human PAH, in TNF-Tg endothelial cells. E. Loss of canonical venous and arterial markers vWF and up-regulation of LYVE1 in arterial /venous endothelial cells. F. Over-expression of adhesion molecules PECAM1 and loss of VCAM1 in endothelial cells. G. Over-expression of basement membrane components HSPG2 and Col4a1 across multiple endothelial cell types
Figure 2. Single cell digestion, sorting, and characterization of endothelial cell phenotypes in TNF-Tg lungs. A. Flow cytometric gating strategy to deplete lungs of CD45+ and CD326+ cells. B. uMAP projection demonstrating 15 pulmonary endothelial and menenchymal cellular populations identified after single cell RNA sequencing across 43,455 cells sequenced. (inset upper) heatmap demonstrating canonical genes used to identify cellular sub-populations C. uMAP projection demonstrating subsets of endothelial cells, note decreased cellular density of general capillary endothelial cells (gCAP) particularly at the 14 and 20 week timepoints in TNF-Tg mice. D. Up-regulation of CD34 and down-regulation of CD74, markers previously shown to be differentially regulated in human PAH, in TNF-Tg endothelial cells. E. Loss of canonical venous and arterial markers vWF and up-regulation of LYVE1 in arterial /venous endothelial cells. F. Over-expression of adhesion molecules PECAM1 and loss of VCAM1 in endothelial cells. G. Over-expression of basement membrane components HSPG2 and Col4a1 across multiple endothelial cell types
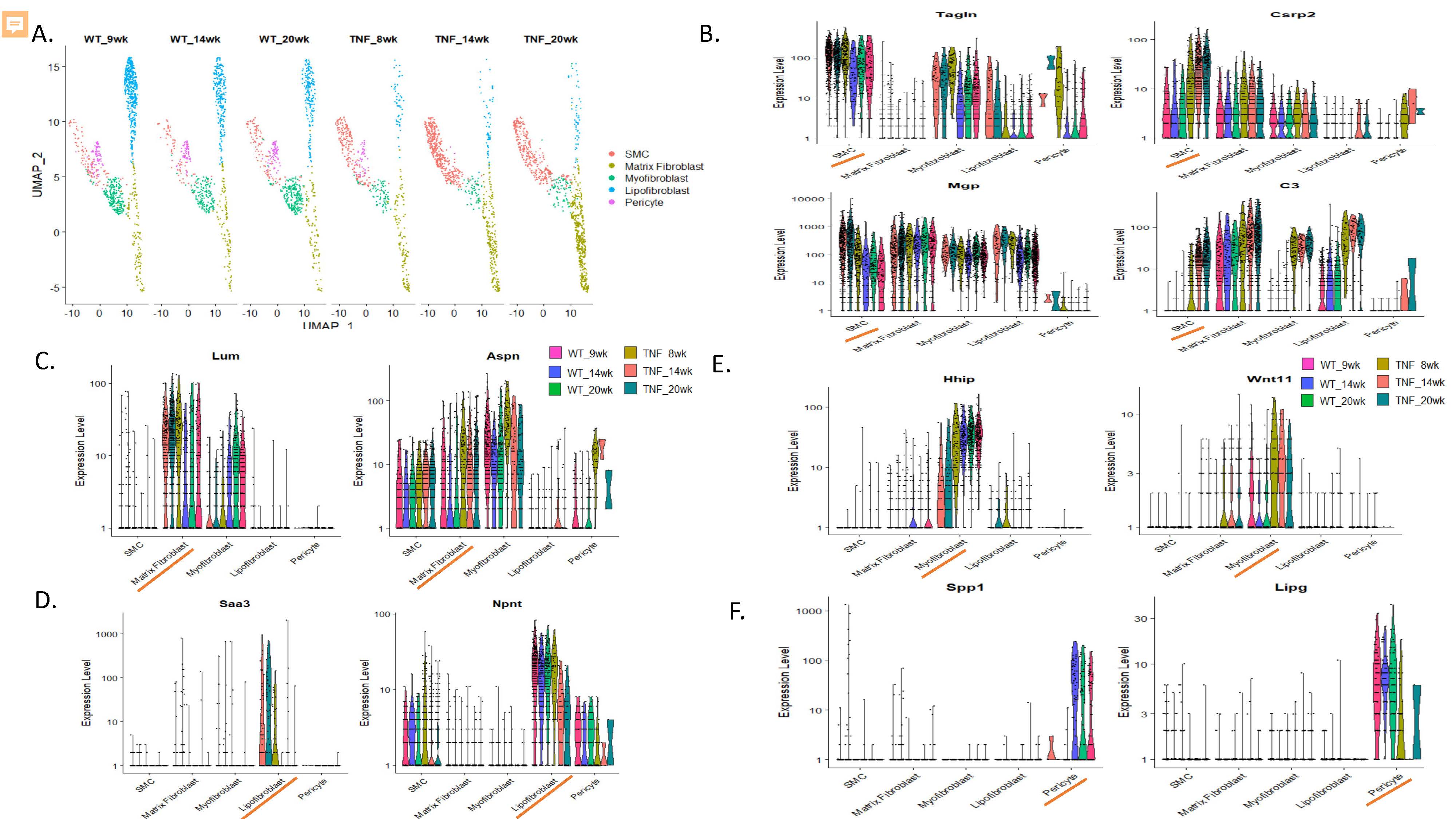 Figure 3. Alterations in mesenchymal cell types in TNF-Tg lungs. A. uMAP projection demonstrating subsets of mesenchymal cells, note increases in smooth muscle cells and matrix fibroblast populations and decreases in pericytes, lipofibroblasts, and myofibroblasts in TNF-Tg mice. C. Over-expression of TAGLN, CSRP2, MGP, and C3 in TNF-Tg smooth muscle cells. D. Over-expression of LUM and DCN in matrix fibroblasts. D. Loss of NPNT and over-expression of SAA3 in TNF-Tg lipofibroblasts. E. Loss of HHIP and over-expression of WNT11 in TNF-Tg myofibroblasts. F. Loss of SPP1 and LIPG expression in TNF-Tg lung pericytes
Figure 3. Alterations in mesenchymal cell types in TNF-Tg lungs. A. uMAP projection demonstrating subsets of mesenchymal cells, note increases in smooth muscle cells and matrix fibroblast populations and decreases in pericytes, lipofibroblasts, and myofibroblasts in TNF-Tg mice. C. Over-expression of TAGLN, CSRP2, MGP, and C3 in TNF-Tg smooth muscle cells. D. Over-expression of LUM and DCN in matrix fibroblasts. D. Loss of NPNT and over-expression of SAA3 in TNF-Tg lipofibroblasts. E. Loss of HHIP and over-expression of WNT11 in TNF-Tg myofibroblasts. F. Loss of SPP1 and LIPG expression in TNF-Tg lung pericytes
Disclosures: S. Duemmel, None; M. Nuzzo, None; S. Bhattacharya, None; Q. Xu, None; A. Mohan, None; B. Korman, None.
Background/Purpose: We previously demonstrated that TNF-transgenic (TNF-Tg) mice have findings consistent with connective-tissue disease associated pulmonary arterial hypertension (CTD-PAH), and that this pathology is mediated by non-hematopoietic cells. Because other TNF-transgenic models have shown divergent TNF receptor pathology between arthritis and visceral disease, we sought to understand the contribution of TNF receptors 1 and 2 (TNFR1 and TNFR2) in PAH pathogenesis. Moreover, to better understand the cellular mechanism contributing to PAH in this model, we sought to evaluate the roles of endothelial and stromal cell alterations in lungs from TNF-Tg mice.
Methods: TNF-Tg mice were crossed with mice deficient in either TNFR1 or TNFR2. Single and double-transgenic mice (5-6 per group) were assessed at 4.5 months of age for pulmonary vascular alterations by histomorphometry and micro-CT angiography, right ventricular pathology by histology, echocardiography, and right heart catheterization. Joint erosion was assessed clinically and by micro-CT. Lungs for WT and TNF-Tg mice (n=3 per timepoint per group) were harvested at 1.5, 3.5, and 5 months of age and were digested to a single cell suspension. Flow cytometry was used to deplete hematopoietic (CD45+) and epithelial (CD326+) cells and single cell RNA sequencing (scRNAseq) was performed on the resulting endothelial and mesenchymal cells. Library preparation was done on enzymatically digested cells (n=45,4335) using a 10X Genomics® Chromium instrument and sequencing was performed on an Illumina NovaSeq6000. ScRNA-seq reads were examined for quality and transcripts were mapped to reference mouse genome and assigned to cells of origin using the TopCell atlas. Analysis was performed using Seurat v3.4 to identify, cluster, and visualize distinct cell populations by PCA and UMAP methodologies and to perform differential gene expression analysis.
Results: TNFR1-KO/TNF-Tg mice were protected from PAH and arthritis by clinical, histologic, imaging, and hemodynamic assessment whereas TNFR2-KO/TNF-Tg mice had disease which was indistinguishable from TNF-Tg mice (Figure 1). Single cell RNA-sequencing of lungs revealed that TNF-Tg mice had reduced endothelial cell numbers, particularly gCAP cells, and that TNF-Tg endothelial cells lost canonical endothelial markers (vWF) but over-expressed genes associated with precursors (CD34), inflammation (CD74), adhesion (VCAM1), and the basement membrane (HSPG2, COL4A1). (Figure 2) Additionally, analysis of stromal cells demonstrated that TNF-Tg mice accumulated smooth muscle cells with a proliferative/invasive phenotype, lost pericytes, and showed shifts in fibroblasts with loss of lipofibrobasts and myofibroblasts and increases in matrix fibroblasts. (Figure 3).
Conclusion: TNFR1 rather than TNFR2 is responsible for TNF mediated pulmonary endothelial and stromal pathology which results in PAH in a mouse model of CTD-PAH. Single cell RNA sequencing revealed important TNF-driven changes in cellular phenotype in the endothelial, smooth muscle, pericyte, and fibroblast lineages.
 Figure 1. TNFR1 mediates PAH and arthritis phenotype in a mouse model of CTD-PAH. A. Lung histology (H&E, 20x) images demonstrate that TNF-Tg and TNFR2-KO/TNF-Tg mice, but not TNFR1-KO/TNF-Tg mice have pulmonary inflammation and vascular occlusion (arrowheads). B. Cardiac histology (H&E, 4x) demonstrates RV hypertrophy in TNFR2-KO/TNF-Tg (**) mice but not TNFR1-KO/TNF-Tg mice (*). C. (Left) Quantification of pulmonary vessel media perimeter (Right) Proportion of partially and fully occluded vessels in each phenotype. D. (Left) RV area measured by histomorphometry, (Middle) RV end diastolic area measured by echocardiography (Right) RV systolic pressure measured by right heart catheterization.
Figure 1. TNFR1 mediates PAH and arthritis phenotype in a mouse model of CTD-PAH. A. Lung histology (H&E, 20x) images demonstrate that TNF-Tg and TNFR2-KO/TNF-Tg mice, but not TNFR1-KO/TNF-Tg mice have pulmonary inflammation and vascular occlusion (arrowheads). B. Cardiac histology (H&E, 4x) demonstrates RV hypertrophy in TNFR2-KO/TNF-Tg (**) mice but not TNFR1-KO/TNF-Tg mice (*). C. (Left) Quantification of pulmonary vessel media perimeter (Right) Proportion of partially and fully occluded vessels in each phenotype. D. (Left) RV area measured by histomorphometry, (Middle) RV end diastolic area measured by echocardiography (Right) RV systolic pressure measured by right heart catheterization.  Figure 2. Single cell digestion, sorting, and characterization of endothelial cell phenotypes in TNF-Tg lungs. A. Flow cytometric gating strategy to deplete lungs of CD45+ and CD326+ cells. B. uMAP projection demonstrating 15 pulmonary endothelial and menenchymal cellular populations identified after single cell RNA sequencing across 43,455 cells sequenced. (inset upper) heatmap demonstrating canonical genes used to identify cellular sub-populations C. uMAP projection demonstrating subsets of endothelial cells, note decreased cellular density of general capillary endothelial cells (gCAP) particularly at the 14 and 20 week timepoints in TNF-Tg mice. D. Up-regulation of CD34 and down-regulation of CD74, markers previously shown to be differentially regulated in human PAH, in TNF-Tg endothelial cells. E. Loss of canonical venous and arterial markers vWF and up-regulation of LYVE1 in arterial /venous endothelial cells. F. Over-expression of adhesion molecules PECAM1 and loss of VCAM1 in endothelial cells. G. Over-expression of basement membrane components HSPG2 and Col4a1 across multiple endothelial cell types
Figure 2. Single cell digestion, sorting, and characterization of endothelial cell phenotypes in TNF-Tg lungs. A. Flow cytometric gating strategy to deplete lungs of CD45+ and CD326+ cells. B. uMAP projection demonstrating 15 pulmonary endothelial and menenchymal cellular populations identified after single cell RNA sequencing across 43,455 cells sequenced. (inset upper) heatmap demonstrating canonical genes used to identify cellular sub-populations C. uMAP projection demonstrating subsets of endothelial cells, note decreased cellular density of general capillary endothelial cells (gCAP) particularly at the 14 and 20 week timepoints in TNF-Tg mice. D. Up-regulation of CD34 and down-regulation of CD74, markers previously shown to be differentially regulated in human PAH, in TNF-Tg endothelial cells. E. Loss of canonical venous and arterial markers vWF and up-regulation of LYVE1 in arterial /venous endothelial cells. F. Over-expression of adhesion molecules PECAM1 and loss of VCAM1 in endothelial cells. G. Over-expression of basement membrane components HSPG2 and Col4a1 across multiple endothelial cell types Figure 3. Alterations in mesenchymal cell types in TNF-Tg lungs. A. uMAP projection demonstrating subsets of mesenchymal cells, note increases in smooth muscle cells and matrix fibroblast populations and decreases in pericytes, lipofibroblasts, and myofibroblasts in TNF-Tg mice. C. Over-expression of TAGLN, CSRP2, MGP, and C3 in TNF-Tg smooth muscle cells. D. Over-expression of LUM and DCN in matrix fibroblasts. D. Loss of NPNT and over-expression of SAA3 in TNF-Tg lipofibroblasts. E. Loss of HHIP and over-expression of WNT11 in TNF-Tg myofibroblasts. F. Loss of SPP1 and LIPG expression in TNF-Tg lung pericytes
Figure 3. Alterations in mesenchymal cell types in TNF-Tg lungs. A. uMAP projection demonstrating subsets of mesenchymal cells, note increases in smooth muscle cells and matrix fibroblast populations and decreases in pericytes, lipofibroblasts, and myofibroblasts in TNF-Tg mice. C. Over-expression of TAGLN, CSRP2, MGP, and C3 in TNF-Tg smooth muscle cells. D. Over-expression of LUM and DCN in matrix fibroblasts. D. Loss of NPNT and over-expression of SAA3 in TNF-Tg lipofibroblasts. E. Loss of HHIP and over-expression of WNT11 in TNF-Tg myofibroblasts. F. Loss of SPP1 and LIPG expression in TNF-Tg lung pericytesDisclosures: S. Duemmel, None; M. Nuzzo, None; S. Bhattacharya, None; Q. Xu, None; A. Mohan, None; B. Korman, None.

