Back
Poster Session A
Osteoarthritis (OA) and related disorders
Session: (0017–0033) Osteoarthritis and Joint Biology – Basic Science Poster
0017: Intra-articular Injection of Bacterial DNA Amplified from Human OA Patient Cartilage Worsens OA Outcomes in Mice
Saturday, November 12, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
- LS
Leoni Schlupp, BS
Oklahoma Medical Research Foundation
Oklahoma City, OK, United States
Abstract Poster Presenter(s)
Leoni Schlupp1, Emmaline Prinz1, Vladislav Izda2, Emily Nguyen1, Christopher Dunn3 and Matlock Jeffries1, 1Oklahoma Medical Research Foundation, Oklahoma City, OK, 2Oklahoma Medical Research Foundation, New York, NY, 3University of Oklahoma Health Sciences Center, Edmond, OK
Background/Purpose: We have previous demonstrated a bacterial DNA signature within cartilage of humans and mice and shown shifts in this signature with OA development. However, whether this bacterial DNA plays a pathogenic role in the development of OA is unclear. In this experiment, evaluated whether purified, amplified bacterial DNA from human OA patients accelerated histological signs of OA following intraarticular injection into germ-free B6 mice.
Methods: Human knee cartilage was obtained from OA-free cadaveric donors (n=12) and eroded sections of end-stage OA patients undergoing joint replacement (n=24). Bacterial DNA was separated from human DNA using an MBD2-Fc magnetic bead approach and amplified using unbiased whole-genome amplification (Qiagen REPLI-g), then cleaned, concentrated, resuspended in buffer adjusted to 0.90 w/v NaCl (normal), and decontaminated. The absence of endotoxin was confirmed using a Pierce chromogenic endotoxin assay. 16s microbiome composition analysis was performed on sample pools pre- and post-amplification. 1000ng of amplified DNA (OA-free or eroded-OA) in 2.5uL, or an equivalent volume of normal saline, were injected unilaterally into a hind knee of C57BL6n germ-free animals (n=4 eroded, n=5 healthy, n=5 saline). 14 days later, mice underwent DMM surgery in the same knee under aseptic conditions within the germ-free facility. Sterility of germ-free isolators were monitored by weekly fecal pellet bacterial plating. Eight weeks later, mice were sacrificed, knee samples fixed, embedded, sectioned, stained, and graded using the OARSI histopathologic scoring system.
Results: Whole-genome amplification did not significantly change bacterial species representation by 16s sequencing. No significant endotoxin was detected in post-amplified specimens (0.018-0.022 EU/mL). No significant differences in mean OARSI score were found comparing saline-injected with healthy human-injected animals post-DMM (1.38±0.67 vs. 1.33±0.54, mean±SEM, P=0.90). A significant increase was found in human OA-injected animals (2.43±0.66) compared to both saline-injected (P=0.03) and healthy-injected (P=0.05). Within the contralateral (non-injected, non-DMM) side, no differences were seen comparing saline and healthy-injected animals (0.22±0.18 vs. 0.21±0.33, respectively, P=0.8); however, a significant increase was seen in human OA-injected animals (1.06±0.10) compared to both saline-injected (P=0.0002) and healthy-injected (P=0.0006) animals (Figure 1). 16S analysis of cartilage samples post-DMM is ongoing.
Conclusion: Bacterial DNA amplified from human OA patients has pro-OA effects when delivered into germ-free animals although no effects were seen when healthy human cartilage-amplified bacterial DNA was delivered; the source of this disparity is unclear although significant differences in bacterial composition exist between these two cartilage sources. Surprisingly, we also noted increases in histopathological score in the contralateral nonoperated side in OA-injected animals, suggesting a systemic effect of intraarticular injection. Future work should focus on confirmation of our findings, expansion to non-germ-free animals, and mechanistic evaluations.
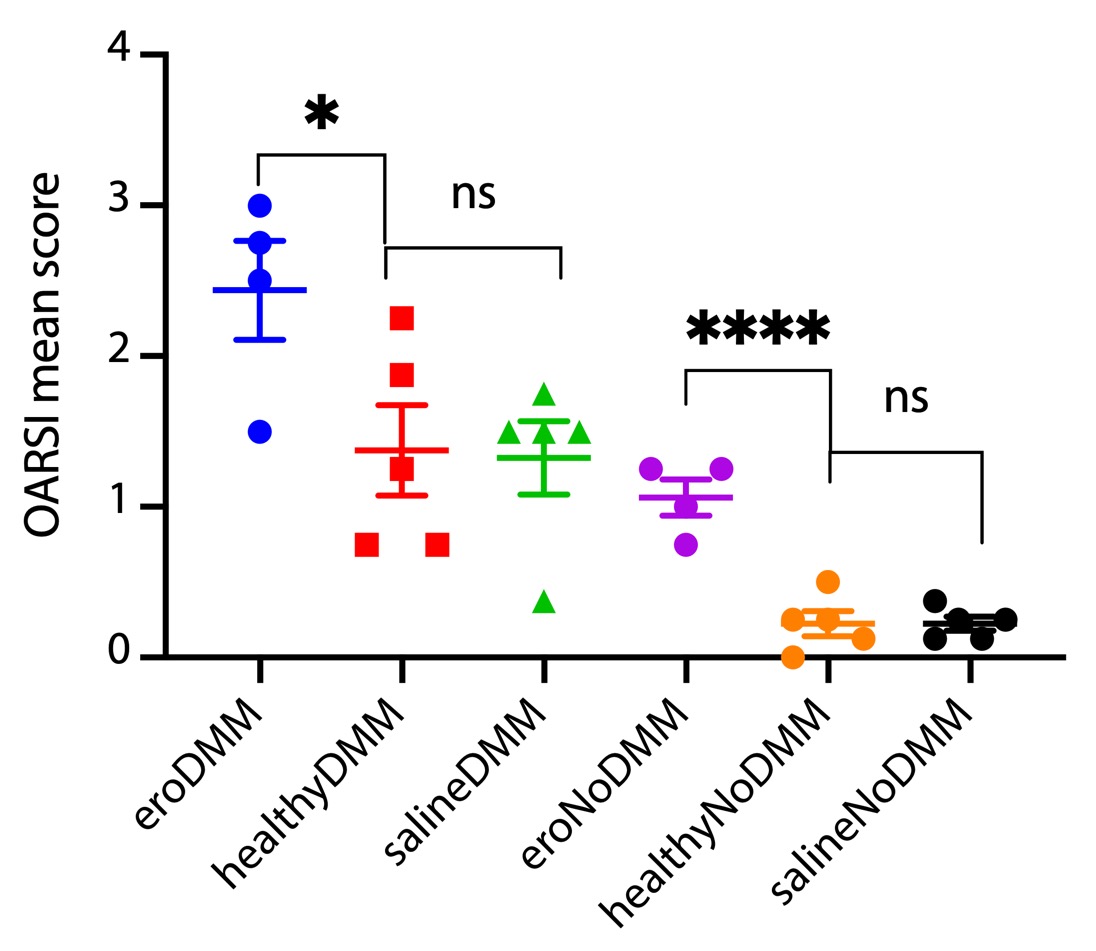 Figure 1: OARSI OA histopathologic scores of knee joints from mice after unilateral joint injection of amplified microbial DNA and DMM, and contralateral non-operated, non-injected knees.
Figure 1: OARSI OA histopathologic scores of knee joints from mice after unilateral joint injection of amplified microbial DNA and DMM, and contralateral non-operated, non-injected knees.
Disclosures: L. Schlupp, None; E. Prinz, None; V. Izda, None; E. Nguyen, None; C. Dunn, None; M. Jeffries, None.
Background/Purpose: We have previous demonstrated a bacterial DNA signature within cartilage of humans and mice and shown shifts in this signature with OA development. However, whether this bacterial DNA plays a pathogenic role in the development of OA is unclear. In this experiment, evaluated whether purified, amplified bacterial DNA from human OA patients accelerated histological signs of OA following intraarticular injection into germ-free B6 mice.
Methods: Human knee cartilage was obtained from OA-free cadaveric donors (n=12) and eroded sections of end-stage OA patients undergoing joint replacement (n=24). Bacterial DNA was separated from human DNA using an MBD2-Fc magnetic bead approach and amplified using unbiased whole-genome amplification (Qiagen REPLI-g), then cleaned, concentrated, resuspended in buffer adjusted to 0.90 w/v NaCl (normal), and decontaminated. The absence of endotoxin was confirmed using a Pierce chromogenic endotoxin assay. 16s microbiome composition analysis was performed on sample pools pre- and post-amplification. 1000ng of amplified DNA (OA-free or eroded-OA) in 2.5uL, or an equivalent volume of normal saline, were injected unilaterally into a hind knee of C57BL6n germ-free animals (n=4 eroded, n=5 healthy, n=5 saline). 14 days later, mice underwent DMM surgery in the same knee under aseptic conditions within the germ-free facility. Sterility of germ-free isolators were monitored by weekly fecal pellet bacterial plating. Eight weeks later, mice were sacrificed, knee samples fixed, embedded, sectioned, stained, and graded using the OARSI histopathologic scoring system.
Results: Whole-genome amplification did not significantly change bacterial species representation by 16s sequencing. No significant endotoxin was detected in post-amplified specimens (0.018-0.022 EU/mL). No significant differences in mean OARSI score were found comparing saline-injected with healthy human-injected animals post-DMM (1.38±0.67 vs. 1.33±0.54, mean±SEM, P=0.90). A significant increase was found in human OA-injected animals (2.43±0.66) compared to both saline-injected (P=0.03) and healthy-injected (P=0.05). Within the contralateral (non-injected, non-DMM) side, no differences were seen comparing saline and healthy-injected animals (0.22±0.18 vs. 0.21±0.33, respectively, P=0.8); however, a significant increase was seen in human OA-injected animals (1.06±0.10) compared to both saline-injected (P=0.0002) and healthy-injected (P=0.0006) animals (Figure 1). 16S analysis of cartilage samples post-DMM is ongoing.
Conclusion: Bacterial DNA amplified from human OA patients has pro-OA effects when delivered into germ-free animals although no effects were seen when healthy human cartilage-amplified bacterial DNA was delivered; the source of this disparity is unclear although significant differences in bacterial composition exist between these two cartilage sources. Surprisingly, we also noted increases in histopathological score in the contralateral nonoperated side in OA-injected animals, suggesting a systemic effect of intraarticular injection. Future work should focus on confirmation of our findings, expansion to non-germ-free animals, and mechanistic evaluations.
 Figure 1: OARSI OA histopathologic scores of knee joints from mice after unilateral joint injection of amplified microbial DNA and DMM, and contralateral non-operated, non-injected knees.
Figure 1: OARSI OA histopathologic scores of knee joints from mice after unilateral joint injection of amplified microbial DNA and DMM, and contralateral non-operated, non-injected knees.Disclosures: L. Schlupp, None; E. Prinz, None; V. Izda, None; E. Nguyen, None; C. Dunn, None; M. Jeffries, None.

