Back
Poster Session C
Rheumatoid arthritis (RA)
Session: (1387–1416) RA – Diagnosis, Manifestations, and Outcomes Poster III
1415: Different Bioactive Lipid Profile Predicts Response to TNF or IL6 Inhibitors in Rheumatoid Arthritis: Result of the CorEvitas CERTAIN Comparative Effectiveness Study
Sunday, November 13, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
- MG
Monica Guma, MD, PhD
UCSD
La Jolla, CA, United States
Abstract Poster Presenter(s)
Mona Alotaibi1, Roxana Coras2, Dimitrios Pappas3, Ted Mikuls4, Joel Kremer5, Geoffrey Thiele6, mohit jain1 and Monica Guma7, 1Department of Medicine, School of Medicine, University of California San Diego, San Diego, CA, 2University of California San Diego, San Diego, CA, 3CorEvitas, LLC, Waltham, MA, 4Division of Rheumatology, University of Nebraska Medical Center, Omaha, NE, 5The Corrona Research Foundation, Delray Beach, FL, 6University of Nebraska Medical Center, Omaha, NE, 7UCSD, La Jolla, CA
Background/Purpose: Circulating bioactive lipids can provide information about the pathogenesis of specific diseases and potentially help predict therapeutic response. Choosing the right biological therapy earlier in the course of rheumatoid arthritis (RA) could help reach the goal of remission. We hypothesized that circulating bioactive lipids at baseline would identify specific metabolic profiles that predict patient response to therapy and would define elements of metabolic pathobiology in arthritis.
Methods: Bioactive lipids were measured in plasma from two cohorts of RA patients from the CorEvitas (formerly known as Corrona) CERTAIN registry at baseline prior to treatment with TNF inhibitors (all biologic naïve, N=102) or anti-IL6 (all previously exposed to biologics, N=114). Response to treatment was categorized by minimal clinically important difference (MCID) in Clinical Disease Activity Index (CDAI) at 6 months after treatment initiation. Patients had to have a 6 month follow up visit. Liquid chromatography (LC) system coupled with high resolution QExactive orbitrap mass spectrometer (LC/MS) was used for bioactive lipids profiling. Around 300 spectral features were identified as potential oxylipins by searching against an in-house MS/MS library. Logistic regression analyses adjusted for gender, age and BMI was perfomed using R software.
Results: 102 patients (average age 54, standard deviation [SD] 12.6, 82% female [83], average BMI 29.7, SD 6.7, average CDAI 27.1, SD 13.7) starting anti-TNF therapy and 114 patients (average age 57, SD 13, 90% female [102], average BMI 30.5, SD 7.4, average CDAI 28.7, SD 13.8) starting tocilizumab were analyzed. Twenty-five oxylipins discriminated between RA patients classified as anti-TNF responders (R, n = 74) and non-responders (NR, n = 28). The anti-inflammatory SPM maresin 2 was increased in R (OR 1.7; 95%CI 1.07–2.9), and oxylipins 15d PGJ2 (OR 0.4; 95%CI 0.2–0.8), and 5,6-diHETE (OR 0.5; 95%CI 0.3–0.9) were decreased in R, among other metabolites Figure 1 and 2. Twenty different metabolites discriminated anti-IL6 R (n=73) and NR (n=41) as shown in Figure 1 and 2. The anti-inflammatory oxylipin 14-15EET (OR 1.7; 95%CI 1.05–2.7) and 8-iso-PGF1a (OR 1.8; 95%CI 1.07–3.1) were increased in R, whereas the pro-inflammatory oxylipins 16-HETE (OR 0.4; 95%CI 0.2–0.9) and 5S-HpETE (OR 0.5; 95%CI 0.26–0.95) were decreased in R.
Conclusion: Circulating bioactive lipid analysis using LC/MS provided a rapid analysis of a wide range of metabolites and can be used to describe metabolic signatures that predict response to therapies. These results lay the groundwork for more deliberate investigations of novel metabolic-based interventions to predict response to therapy and reduce arthritis morbidity.
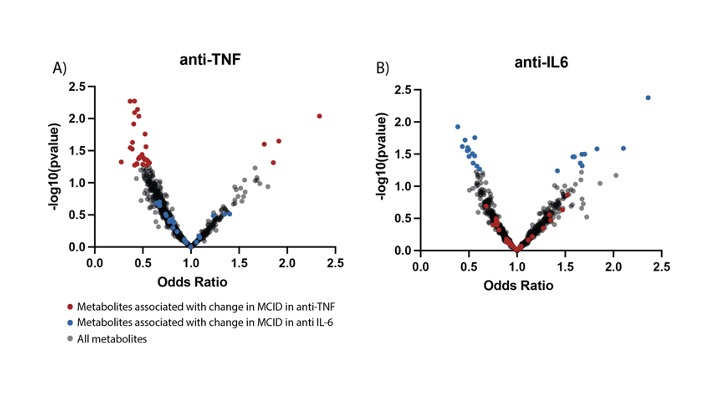 Figure 1.Volcano plots visualizing baseline metabolites associated with responders vs. non responders in a) anti-TNF and b) anti IL-6 therapy groups. Results are derived from multivariate logistic regression analysis of baseline metabolites and response to treatment categorized by MCID. Data plotted as the metabolite against its statistical significance, respectively reported as odds ratio (OR) and -log10(pvalue)
Figure 1.Volcano plots visualizing baseline metabolites associated with responders vs. non responders in a) anti-TNF and b) anti IL-6 therapy groups. Results are derived from multivariate logistic regression analysis of baseline metabolites and response to treatment categorized by MCID. Data plotted as the metabolite against its statistical significance, respectively reported as odds ratio (OR) and -log10(pvalue)
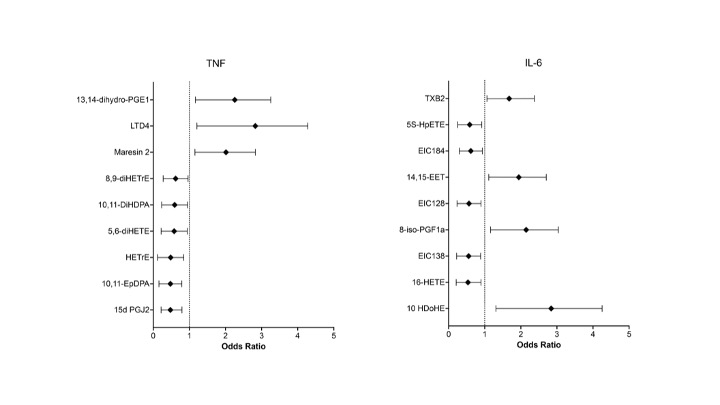 Figure 2. Forest plot visualizing baseline metabolites associated with responders vs. non responders in anti-TNF and anti-IL6 therapy groups. Results are derived from multivariate logistic regression analysis of baseline metabolites and response to treatment categorized by MCID in CDAI
Figure 2. Forest plot visualizing baseline metabolites associated with responders vs. non responders in anti-TNF and anti-IL6 therapy groups. Results are derived from multivariate logistic regression analysis of baseline metabolites and response to treatment categorized by MCID in CDAI
Disclosures: M. Alotaibi, None; R. Coras, None; D. Pappas, CorEvitas, Novartis, Sanofi, Genentech, Roche, AbbVie; T. Mikuls, Gilead Sciences, Bristol-Myers Squibb, Horizon, Sanofi, Pfizer Inc; J. Kremer, CorEvitas; G. Thiele, None; m. jain, Sapient; M. Guma, Pfizer, Novartis, Gilead, Sonoma Bio, Genentech.
Background/Purpose: Circulating bioactive lipids can provide information about the pathogenesis of specific diseases and potentially help predict therapeutic response. Choosing the right biological therapy earlier in the course of rheumatoid arthritis (RA) could help reach the goal of remission. We hypothesized that circulating bioactive lipids at baseline would identify specific metabolic profiles that predict patient response to therapy and would define elements of metabolic pathobiology in arthritis.
Methods: Bioactive lipids were measured in plasma from two cohorts of RA patients from the CorEvitas (formerly known as Corrona) CERTAIN registry at baseline prior to treatment with TNF inhibitors (all biologic naïve, N=102) or anti-IL6 (all previously exposed to biologics, N=114). Response to treatment was categorized by minimal clinically important difference (MCID) in Clinical Disease Activity Index (CDAI) at 6 months after treatment initiation. Patients had to have a 6 month follow up visit. Liquid chromatography (LC) system coupled with high resolution QExactive orbitrap mass spectrometer (LC/MS) was used for bioactive lipids profiling. Around 300 spectral features were identified as potential oxylipins by searching against an in-house MS/MS library. Logistic regression analyses adjusted for gender, age and BMI was perfomed using R software.
Results: 102 patients (average age 54, standard deviation [SD] 12.6, 82% female [83], average BMI 29.7, SD 6.7, average CDAI 27.1, SD 13.7) starting anti-TNF therapy and 114 patients (average age 57, SD 13, 90% female [102], average BMI 30.5, SD 7.4, average CDAI 28.7, SD 13.8) starting tocilizumab were analyzed. Twenty-five oxylipins discriminated between RA patients classified as anti-TNF responders (R, n = 74) and non-responders (NR, n = 28). The anti-inflammatory SPM maresin 2 was increased in R (OR 1.7; 95%CI 1.07–2.9), and oxylipins 15d PGJ2 (OR 0.4; 95%CI 0.2–0.8), and 5,6-diHETE (OR 0.5; 95%CI 0.3–0.9) were decreased in R, among other metabolites Figure 1 and 2. Twenty different metabolites discriminated anti-IL6 R (n=73) and NR (n=41) as shown in Figure 1 and 2. The anti-inflammatory oxylipin 14-15EET (OR 1.7; 95%CI 1.05–2.7) and 8-iso-PGF1a (OR 1.8; 95%CI 1.07–3.1) were increased in R, whereas the pro-inflammatory oxylipins 16-HETE (OR 0.4; 95%CI 0.2–0.9) and 5S-HpETE (OR 0.5; 95%CI 0.26–0.95) were decreased in R.
Conclusion: Circulating bioactive lipid analysis using LC/MS provided a rapid analysis of a wide range of metabolites and can be used to describe metabolic signatures that predict response to therapies. These results lay the groundwork for more deliberate investigations of novel metabolic-based interventions to predict response to therapy and reduce arthritis morbidity.
 Figure 1.Volcano plots visualizing baseline metabolites associated with responders vs. non responders in a) anti-TNF and b) anti IL-6 therapy groups. Results are derived from multivariate logistic regression analysis of baseline metabolites and response to treatment categorized by MCID. Data plotted as the metabolite against its statistical significance, respectively reported as odds ratio (OR) and -log10(pvalue)
Figure 1.Volcano plots visualizing baseline metabolites associated with responders vs. non responders in a) anti-TNF and b) anti IL-6 therapy groups. Results are derived from multivariate logistic regression analysis of baseline metabolites and response to treatment categorized by MCID. Data plotted as the metabolite against its statistical significance, respectively reported as odds ratio (OR) and -log10(pvalue) Figure 2. Forest plot visualizing baseline metabolites associated with responders vs. non responders in anti-TNF and anti-IL6 therapy groups. Results are derived from multivariate logistic regression analysis of baseline metabolites and response to treatment categorized by MCID in CDAI
Figure 2. Forest plot visualizing baseline metabolites associated with responders vs. non responders in anti-TNF and anti-IL6 therapy groups. Results are derived from multivariate logistic regression analysis of baseline metabolites and response to treatment categorized by MCID in CDAIDisclosures: M. Alotaibi, None; R. Coras, None; D. Pappas, CorEvitas, Novartis, Sanofi, Genentech, Roche, AbbVie; T. Mikuls, Gilead Sciences, Bristol-Myers Squibb, Horizon, Sanofi, Pfizer Inc; J. Kremer, CorEvitas; G. Thiele, None; m. jain, Sapient; M. Guma, Pfizer, Novartis, Gilead, Sonoma Bio, Genentech.

