Back
Poster Session A
Systemic lupus erythematosus (SLE)
Session: (0343–0371) SLE – Treatment Poster I
0361: Gene Expression Pathways Modulated by Anifrolumab in Systemic Lupus Erythematosus
Saturday, November 12, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
- TB
Tina Baker, PhD
Astrazeneca
Cambridge, United Kingdom
Abstract Poster Presenter(s)
Tina Baker1, Sharifian Hoda2, Paul Newcombe1, Mark Lazarus1, Madhu Ramaswamy3, Nicola Ferrari1, Daniel Muthas2, Hussein Al-Mossawi1, Raj Tummala3, Eric F. Morand4, Richard A. Furie5, Edward Vital6 and Philip Brohawn3, 1AstraZeneca, Cambridge, United Kingdom, 2AstraZeneca, Gothenburg, Sweden, 3AstraZeneca, Gaithersburg, MD, 4Monash University, Melbourne, Australia, 5Northwell Health, Great Neck, NY, 6University of Leeds, Leeds, England, United Kingdom
Background/Purpose: Anifrolumab is a monoclonal antibody to IFN-α receptor 1 that inhibits type I IFN signaling. In the phase 3 TULIP-1 and TULIP-2 trials, anifrolumab resulted in a decreased type I IFN gene signature and favorable clinical outcomes compared to standard of care (SOC) in adult patients with moderate to severe SLE.1-3 The current analysis sought to identify biomarkers associated with anifrolumab treatment using transcriptomic assessments of longitudinal whole blood samples from these trials.
Methods: All patients fulfilled the 1997 ACR criteria for SLE. Whole blood was collected at baseline, Week 24, and Week 52 from 502 patients receiving anifrolumab (300 mg) or placebo plus SOC and were assessed for transcriptomics using genome-wide RNA sequencing. Low count genes (< 5 counts in >50% of the samples) were excluded from differential expression analysis. A longitudinal mixed-effect model assessed if individual genes were statistically different between anifrolumab and placebo groups at Weeks 24 and 52 from baseline. The model was corrected for baseline SLEDAI-2K (≥10 points) and oral glucocorticoids dose (≥10 mg/day) as stratification factors. Multiple testing correction was performed using the Benjamini–Hochberg False Discovery Rate (FDR) procedure. Week 52 results were used in the pathway analysis using Gene Set Enrichment Analysis and MetaBase pathway database, in addition to the blood modules membership analysis.4
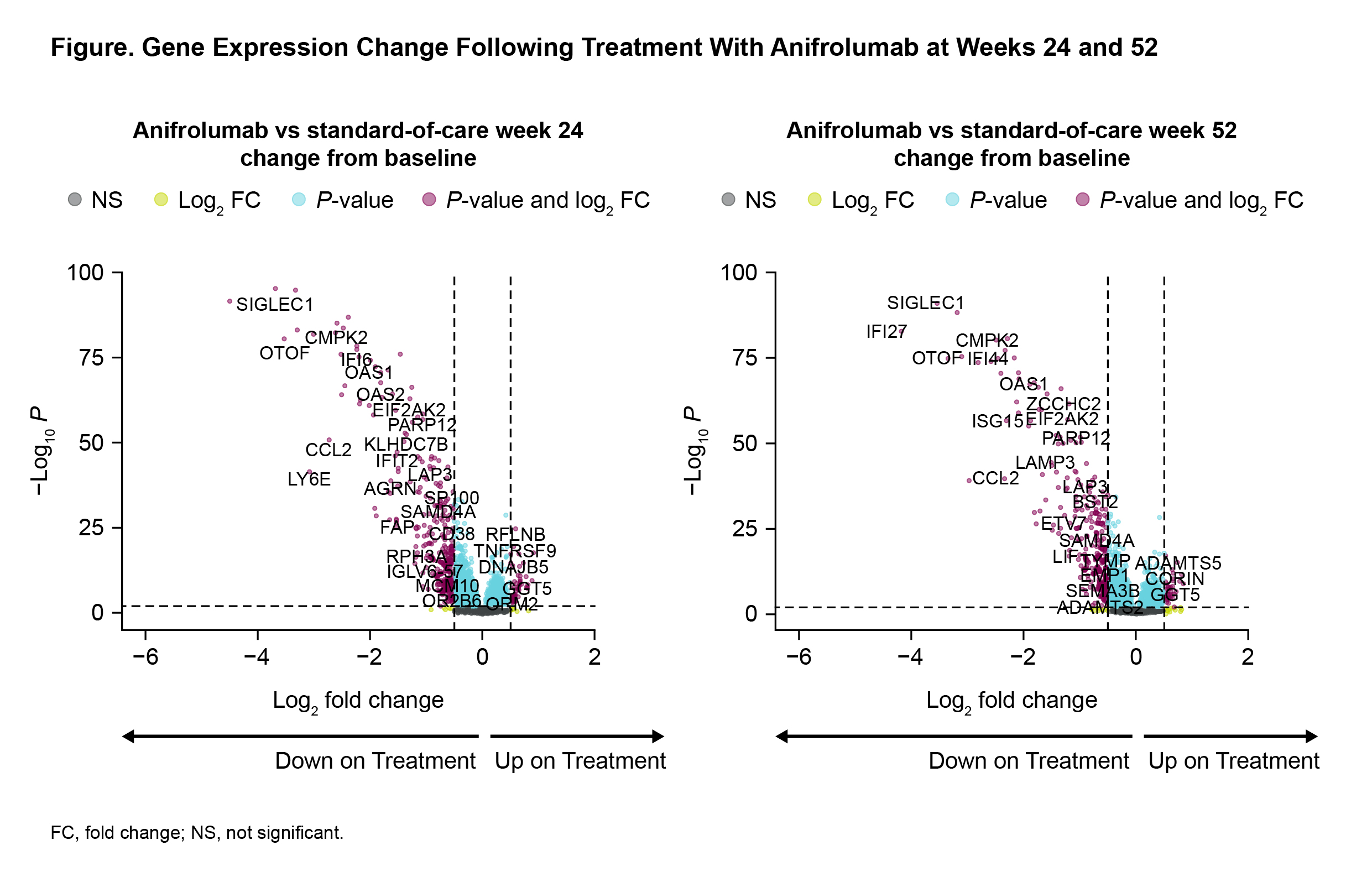
Results: Longitudinal analysis of transcriptomic data identified 2780 and 3232 genes that were differentially impacted by anifrolumab vs placebo at Weeks 24 and 52, respectively. Of those, 2079 genes were modulated at both timepoints (FDR < 0.05). Several IFN-associated genes, including SIGLEC1, IFI44, IFI44L, IFIT1, MX1, and LAMP3, were significantly downregulated at Week 24 (Figure). Pathway analysis revealed upregulated pathways related to protein translation and synthesis, as well as multiple type I IFN and immune response mechanisms (FDR < 0.01). Importantly, common pathways annotated "NETosis in SLE" and "dysregulation of germinal center response" were among those most downregulated by anifrolumab. Blood module analysis supported these findings by showing strong downregulation of the IFN and plasma cell modules, and upregulation of protein synthesis.
Conclusion: Our findings provide mechanistic insights into how type I IFN blockade by anifrolumab affects downstream pathways that are important in SLE.
References:
1. Furie R, et al. Arthritis Rheumatol. 2017;69:376–86.
2. Furie RA, et al. Lancet Rheumatol. 2019;1:e208–19.
3. Morand EF, et al. N Engl J Med. 2020;382:211–21.
4. Altman MC, et al. Nat Commun. 2021;12:4385.
Disclosures: T. Baker, AstraZeneca; S. Hoda, AstraZeneca; P. Newcombe, AstraZeneca; M. Lazarus, AstraZeneca, Novartis; M. Ramaswamy, AstraZeneca; N. Ferrari, AstraZeneca; D. Muthas, AstraZeneca; H. Al-Mossawi, AstraZeneca; R. Tummala, AstraZeneca; E. Morand, AbbVie, Amgen, AstraZeneca, Biogen, BristolMyersSquibb, Eli Lilly, Genentech, GlaxoSmithKline, Janssen, Novartis, Servier, UCB, EMD Soreno; R. Furie, AstraZeneca, Biogen; E. Vital, AstraZeneca; P. Brohawn, AstraZeneca.
Background/Purpose: Anifrolumab is a monoclonal antibody to IFN-α receptor 1 that inhibits type I IFN signaling. In the phase 3 TULIP-1 and TULIP-2 trials, anifrolumab resulted in a decreased type I IFN gene signature and favorable clinical outcomes compared to standard of care (SOC) in adult patients with moderate to severe SLE.1-3 The current analysis sought to identify biomarkers associated with anifrolumab treatment using transcriptomic assessments of longitudinal whole blood samples from these trials.
Methods: All patients fulfilled the 1997 ACR criteria for SLE. Whole blood was collected at baseline, Week 24, and Week 52 from 502 patients receiving anifrolumab (300 mg) or placebo plus SOC and were assessed for transcriptomics using genome-wide RNA sequencing. Low count genes (< 5 counts in >50% of the samples) were excluded from differential expression analysis. A longitudinal mixed-effect model assessed if individual genes were statistically different between anifrolumab and placebo groups at Weeks 24 and 52 from baseline. The model was corrected for baseline SLEDAI-2K (≥10 points) and oral glucocorticoids dose (≥10 mg/day) as stratification factors. Multiple testing correction was performed using the Benjamini–Hochberg False Discovery Rate (FDR) procedure. Week 52 results were used in the pathway analysis using Gene Set Enrichment Analysis and MetaBase pathway database, in addition to the blood modules membership analysis.4
Results: Longitudinal analysis of transcriptomic data identified 2780 and 3232 genes that were differentially impacted by anifrolumab vs placebo at Weeks 24 and 52, respectively. Of those, 2079 genes were modulated at both timepoints (FDR < 0.05). Several IFN-associated genes, including SIGLEC1, IFI44, IFI44L, IFIT1, MX1, and LAMP3, were significantly downregulated at Week 24 (Figure). Pathway analysis revealed upregulated pathways related to protein translation and synthesis, as well as multiple type I IFN and immune response mechanisms (FDR < 0.01). Importantly, common pathways annotated "NETosis in SLE" and "dysregulation of germinal center response" were among those most downregulated by anifrolumab. Blood module analysis supported these findings by showing strong downregulation of the IFN and plasma cell modules, and upregulation of protein synthesis.
Conclusion: Our findings provide mechanistic insights into how type I IFN blockade by anifrolumab affects downstream pathways that are important in SLE.
References:
1. Furie R, et al. Arthritis Rheumatol. 2017;69:376–86.
2. Furie RA, et al. Lancet Rheumatol. 2019;1:e208–19.
3. Morand EF, et al. N Engl J Med. 2020;382:211–21.
4. Altman MC, et al. Nat Commun. 2021;12:4385.

Disclosures: T. Baker, AstraZeneca; S. Hoda, AstraZeneca; P. Newcombe, AstraZeneca; M. Lazarus, AstraZeneca, Novartis; M. Ramaswamy, AstraZeneca; N. Ferrari, AstraZeneca; D. Muthas, AstraZeneca; H. Al-Mossawi, AstraZeneca; R. Tummala, AstraZeneca; E. Morand, AbbVie, Amgen, AstraZeneca, Biogen, BristolMyersSquibb, Eli Lilly, Genentech, GlaxoSmithKline, Janssen, Novartis, Servier, UCB, EMD Soreno; R. Furie, AstraZeneca, Biogen; E. Vital, AstraZeneca; P. Brohawn, AstraZeneca.

