Back
Poster Session A
Spondyloarthritis (SpA) including psoriatic arthritis (PsA)
Session: (0372–0402) Spondyloarthritis Including PsA – Diagnosis, Manifestations, and Outcomes Poster I
0386: Quantitative Proteomic Screening Uncovers Diagnostic and Monitoring Serum Biomarkers of Ankylosing Spondylitis
Saturday, November 12, 2022
1:00 PM – 3:00 PM Eastern Time
Location: Virtual Poster Hall
- MH
Mark C. Hwang, MD, MS
McGovern Medical School at the University of Texas Health Science Center at Houston
Houston, TX, United States
Abstract Poster Presenter(s)
Mark Hwang1, Jessica Castillo2, kamala Vanarsa2, Jim Zheng3, Shervin Assassi4, Chandra Mohan2 and John Reveille5, 1McGovern Medical School at the University of Texas Health Science Center at Houston, Houston, TX, 2University of Houston, Houston, TX, 3UTHealth School of Bioinformatics, Houston, TX, 4McGovern Medical School, University of Texas, Houston, TX, 5University of Texas McGovern Medical School, Houston, TX
Background/Purpose: We sought to discover novel serum biomarkers of ankylosing spondylitis (AS) for diagnosis and monitoring disease activity.
Methods: We used biologic-treatment naïve AS and healthy control (HC) patient sera from the Prospective Study of Outcome in Ankylosing Spondylitis UTHealth biorepository. We studied age, gender and race-matched patient and control sera in a 1:1:1 ratio for active, inactive, and HC for a total of 80 patients. Active disease was defined by an Ankylosing Spondylitis Disease Activity Score (ASDAS) ≥2.1 and inactive disease as ASDAS < 1.3. Serum samples were immediately stored < -70°C and not thawed for other experiments. We used the 1320 protein SOMAscan™ assay (SomaLogic; Boulder, CO), which is a sensitive, quantitative, aptameric protein biomarker discovery platform.
T-tests tests were performed for high and low-disease activity AS patients compared to health controls (Diagnosis Analysis) and high compared to low disease activity (Monitoring Analysis) in a 2:1 and 1:1 ratio, respectively. In our analyses, p-values and q-values (false discovery-rate [FDR] corrected p-value) were calculated for all proteins- a p< 0.05 were considered differentially expressed proteins (DEPs).
We used the STRING (https://string-db.org) database to analyze DEPs in our Diagnosis and Monitoring analyses for protein-protein interactions (PPI) using Cytoscape software. The Cytoscape Molecular Complex Detection (MCODE) plugin was used to find clusters in PPI networks, confidence cutoff of 0.4. Lasso regression analysis was performed of our Diagnosis DEPs to determine optimal protein combination in a 1:1 training/test set split cohort of our 80 patients in R.
Results: Of the 1317 proteins detected in our Diagnosis and Monitoring analyses, 367 and 167 (317 and 59, FDR-corrected q< .05) DEPs were detected, respectively (Figure 1). The top 10 up-regulated and down-regulated DEPs based on fold change for diagnostic and monitoring biomarkers are presented in Table 1.
MCODE analyses identified complement regulation/signal transduction, IL-10 signaling/Immune system and Immune system/Interleukin signaling as the top 3 clusters among the diagnostic biomarkers. Complement, Extracellular matrix organization/proteoglycans and MAPK/RAS signaling were the top 3 clusters among the monitoring biomarkers (Figure 2).
Lasso regression identified a Diagnostic 13-protein model (IgA, C5a, SARP-2, SLPI, Cathepsin A, NXPH1, CXCL16, gp130, C4b, Cofilin-1, CHL1, SLAF6 and Macrophage mannose receptor) predictive of AS. This model had a sensitivity of 0.75, specificity of 0.90, a kappa of 0.59, and an overall accuracy of 0.80 (95% CI: 0.61-0.92). The predictive probability of our model to discriminate AS patients vs controls based on ROC curve (95% CI) was 0.79 (0.61-0.96).
Conclusion: We identified multiple candidate AS diagnostic and disease activity monitoring serum biomarkers using a comprehensive proteomic screen. Enrichment analysis identified key pathways in AS diagnosis and monitoring. Lasso regression identified candidate diagnostic serum biomarker panels with modest predictive ability. Further work is required to validate these findings.
<img src=https://www.abstractscorecard.com/uploads/Tasks/upload/17574/QHOPTGBB-1287997-1-ANY.jpg width=440 height=193.300384404174 border=0 style=border-style: none;>
.jpg)
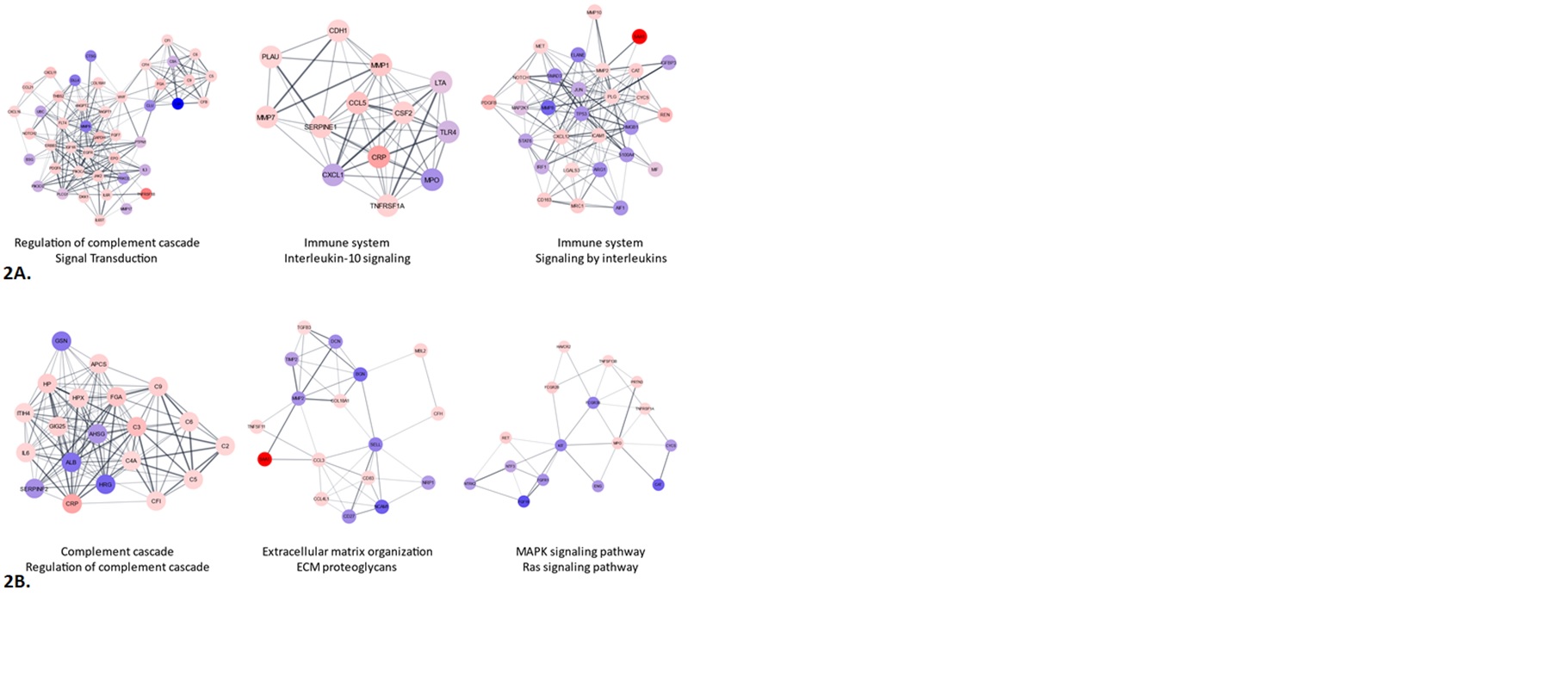 Figure 2. Protein-protein interaction networks 2A. Top 367 Diagnosis DEPs and 2B. Top 157 Monitoring DEPs (T-test p < 0.05) through the Cytoscape string App with a confidence cutoff of 0.4. MCODE clustering was performed and displayed are the top three clusters. The color of each node corresponds to the fold change. Nodes with a fold change less than one range in color from blue to purple while those with a fold change greater than one range from pink to red. The confidence score of each interaction is displayed as the edge thickness and opacity. The top two reactome pathways associated with each cluster are displayed below each cluster.
Figure 2. Protein-protein interaction networks 2A. Top 367 Diagnosis DEPs and 2B. Top 157 Monitoring DEPs (T-test p < 0.05) through the Cytoscape string App with a confidence cutoff of 0.4. MCODE clustering was performed and displayed are the top three clusters. The color of each node corresponds to the fold change. Nodes with a fold change less than one range in color from blue to purple while those with a fold change greater than one range from pink to red. The confidence score of each interaction is displayed as the edge thickness and opacity. The top two reactome pathways associated with each cluster are displayed below each cluster.
Disclosures: M. Hwang, Novartis; J. Castillo, None; k. Vanarsa, None; J. Zheng, None; S. Assassi, Boehringer-Ingelheim, Janssen, Novartis, AstraZeneca, CSL Behring, AbbVie/Abbott; C. Mohan, None; J. Reveille, Eli Lilly.
Background/Purpose: We sought to discover novel serum biomarkers of ankylosing spondylitis (AS) for diagnosis and monitoring disease activity.
Methods: We used biologic-treatment naïve AS and healthy control (HC) patient sera from the Prospective Study of Outcome in Ankylosing Spondylitis UTHealth biorepository. We studied age, gender and race-matched patient and control sera in a 1:1:1 ratio for active, inactive, and HC for a total of 80 patients. Active disease was defined by an Ankylosing Spondylitis Disease Activity Score (ASDAS) ≥2.1 and inactive disease as ASDAS < 1.3. Serum samples were immediately stored < -70°C and not thawed for other experiments. We used the 1320 protein SOMAscan™ assay (SomaLogic; Boulder, CO), which is a sensitive, quantitative, aptameric protein biomarker discovery platform.
T-tests tests were performed for high and low-disease activity AS patients compared to health controls (Diagnosis Analysis) and high compared to low disease activity (Monitoring Analysis) in a 2:1 and 1:1 ratio, respectively. In our analyses, p-values and q-values (false discovery-rate [FDR] corrected p-value) were calculated for all proteins- a p< 0.05 were considered differentially expressed proteins (DEPs).
We used the STRING (https://string-db.org) database to analyze DEPs in our Diagnosis and Monitoring analyses for protein-protein interactions (PPI) using Cytoscape software. The Cytoscape Molecular Complex Detection (MCODE) plugin was used to find clusters in PPI networks, confidence cutoff of 0.4. Lasso regression analysis was performed of our Diagnosis DEPs to determine optimal protein combination in a 1:1 training/test set split cohort of our 80 patients in R.
Results: Of the 1317 proteins detected in our Diagnosis and Monitoring analyses, 367 and 167 (317 and 59, FDR-corrected q< .05) DEPs were detected, respectively (Figure 1). The top 10 up-regulated and down-regulated DEPs based on fold change for diagnostic and monitoring biomarkers are presented in Table 1.
MCODE analyses identified complement regulation/signal transduction, IL-10 signaling/Immune system and Immune system/Interleukin signaling as the top 3 clusters among the diagnostic biomarkers. Complement, Extracellular matrix organization/proteoglycans and MAPK/RAS signaling were the top 3 clusters among the monitoring biomarkers (Figure 2).
Lasso regression identified a Diagnostic 13-protein model (IgA, C5a, SARP-2, SLPI, Cathepsin A, NXPH1, CXCL16, gp130, C4b, Cofilin-1, CHL1, SLAF6 and Macrophage mannose receptor) predictive of AS. This model had a sensitivity of 0.75, specificity of 0.90, a kappa of 0.59, and an overall accuracy of 0.80 (95% CI: 0.61-0.92). The predictive probability of our model to discriminate AS patients vs controls based on ROC curve (95% CI) was 0.79 (0.61-0.96).
Conclusion: We identified multiple candidate AS diagnostic and disease activity monitoring serum biomarkers using a comprehensive proteomic screen. Enrichment analysis identified key pathways in AS diagnosis and monitoring. Lasso regression identified candidate diagnostic serum biomarker panels with modest predictive ability. Further work is required to validate these findings.
<img src=https://www.abstractscorecard.com/uploads/Tasks/upload/17574/QHOPTGBB-1287997-1-ANY.jpg width=440 height=193.300384404174 border=0 style=border-style: none;>
Figure 1. Volcano Plot of all serum Proteins for A. Diagnosis (AS patients vs. Healthy Controls) B. Monitoring (AS High vs. Low Disease Activity patients). Fold change of AS patients compared to controls < 0.5, 0.5-2.0, and >2 are presented in yellow, black, and red, respectively. 367 and 157 proteins were differently expressed for Diagnosis and Monitoring, respectively.
.jpg)
 Figure 2. Protein-protein interaction networks 2A. Top 367 Diagnosis DEPs and 2B. Top 157 Monitoring DEPs (T-test p < 0.05) through the Cytoscape string App with a confidence cutoff of 0.4. MCODE clustering was performed and displayed are the top three clusters. The color of each node corresponds to the fold change. Nodes with a fold change less than one range in color from blue to purple while those with a fold change greater than one range from pink to red. The confidence score of each interaction is displayed as the edge thickness and opacity. The top two reactome pathways associated with each cluster are displayed below each cluster.
Figure 2. Protein-protein interaction networks 2A. Top 367 Diagnosis DEPs and 2B. Top 157 Monitoring DEPs (T-test p < 0.05) through the Cytoscape string App with a confidence cutoff of 0.4. MCODE clustering was performed and displayed are the top three clusters. The color of each node corresponds to the fold change. Nodes with a fold change less than one range in color from blue to purple while those with a fold change greater than one range from pink to red. The confidence score of each interaction is displayed as the edge thickness and opacity. The top two reactome pathways associated with each cluster are displayed below each cluster.Disclosures: M. Hwang, Novartis; J. Castillo, None; k. Vanarsa, None; J. Zheng, None; S. Assassi, Boehringer-Ingelheim, Janssen, Novartis, AstraZeneca, CSL Behring, AbbVie/Abbott; C. Mohan, None; J. Reveille, Eli Lilly.

