Back
Poster Session B
Antiphospholipid Syndrome
Session: (0671–0694) Antiphospholipid Syndrome Poster
0689: Type I Interferon Stimulated Genes Identify Different Phenotypes of Antiphospholipid Syndrome Patients
Sunday, November 13, 2022
9:00 AM – 10:30 AM Eastern Time
Location: Virtual Poster Hall
- SS
Savino Sciascia, MD
University of Turin
Torino, Turin, Italy
Abstract Poster Presenter(s)
Irene Cecchi1, Massimo Radin2, Alice Barinotti2, Silvia Grazietta Foddai2, Elena Rubini2, Ana Suárez3, Simone Baldovino2, Elisa Menegatti2, Dario Roccatello2, Savino Sciascia1 and Javier Rodríguez-Carrio4, 1University of Turin, Torino, Italy, 2University of Turin, Turin, Italy, 3University of Oviedo, Oviedo, Spain, 4University of Oviedo,, Oviedo, Spain
Background/Purpose: At present, a limited number of evidencesis available on the specific role of Type I Interferons (IFN) activation in antiphospholipid antibodies (aPL) positive patients, including aPL carriers, primary antiphospholipid syndrome (PAPS) and APS subjects who presented with an associated autoimmune disease (secondary APS, SAPS), such as systemic lupus erythematosus (SLE).
The aim of this study was to evaluate the differential expression of IFN stimulated genes (ISG) among different subsets of aPL positive subjects.
Methods: A total of 112 patients attending the San Giovanni Bosco Hospital (Turin, Italy) were enrolled, including 31 PAPS, 25 SAPS, 27SLE patients without aPL, 29 aPLcarriers (mean age 48.3±13.3 years, 76% female). Nineteen subjects were also recruited as healthy controls (HCs). Gene expression was evaluated by RT-PCR for the following genes: IFI6, IFI44, IFI44L, MX1,IFI27, OAS1 and RSAD2. Normalized gene expression levels (Z-scores) were averaged into a global IFN signature (IFN score). Differences were measured by Kruskal-Wallis tests and associations among genes were studied by cluster and correspondence analyses. Correlations among genes were plotted by network analyses.
Results: An overall activation of ISG was noted across APS subsets, but differences were noted among genes. Whereas some ISG were already upregulated in the aPL positive group compared to HCs (IFI44, IFI44L, MX1, IFI27, OAS1 and RSAD2, all p< 0.050), other ISG were only in increased SLE (IFI6), MX1 differed between SLE and SAPS, and IFI27 and OAS1 showed differences between PAPS and SAPS. The composite IFN score revealed quantitative differences in the IFN pathway activation across APS subsets, being elevated in aPL carriers/PAPS groups compared to HCs (both p< 0.050) and increasing in SAPS (p< 0.010) and SLE (p< 0.001) groups. Network analyses (Figure 1A) revealed qualitative differences in the gene-gene correlation networks: (i) weaker structures were found in HCs and aPL carriers, compared to stronger and higher-degree networks in SAPS and SLE groups; and (ii) the influence of each node was different across groups. Unsupervised cluster analysis identified 3 clusters (I to III) based on ISG patterns (figure 1B). Clusters usage differed among APS subsets, thus correlating clinical status (Figure 1C). Distinct groups of ISG positively correlate to aPS/PT IgG titre in aPL carriers and PAPS groups (all rho >0.500), whereas no associations were retrieved in SAPS or SLE. No associations with thrombotic events were observed, although IFN composite score and several ISG correlate with the number of thrombotic recurrences under anticoagulation (all rho >0.400).
Conclusion: An overall IFN pathway activation has been observed in aPL positive patients and across all APS subsets. Qualitative and quantitative differences across the APS spectrum can be identified, leading tothe identification of distinct IFN signatures with different clinical value.
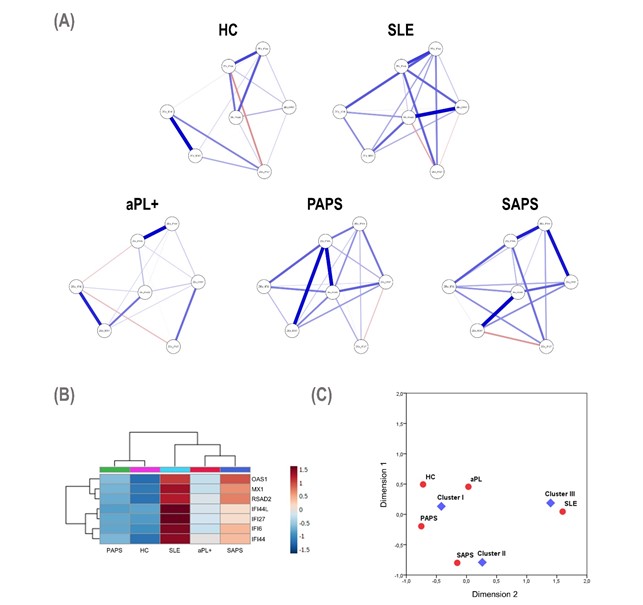 Figure 1.
Figure 1.
1A) Network analyses of the interferon regulated genes among different subsets; 1B and 1C) results of cluster analysis and correlations with different clinical status.
Disclosures: I. Cecchi, None; M. Radin, None; A. Barinotti, None; S. Foddai, None; E. Rubini, None; A. Suárez, None; S. Baldovino, None; E. Menegatti, None; D. Roccatello, None; S. Sciascia, None; J. Rodríguez-Carrio, None.
Background/Purpose: At present, a limited number of evidencesis available on the specific role of Type I Interferons (IFN) activation in antiphospholipid antibodies (aPL) positive patients, including aPL carriers, primary antiphospholipid syndrome (PAPS) and APS subjects who presented with an associated autoimmune disease (secondary APS, SAPS), such as systemic lupus erythematosus (SLE).
The aim of this study was to evaluate the differential expression of IFN stimulated genes (ISG) among different subsets of aPL positive subjects.
Methods: A total of 112 patients attending the San Giovanni Bosco Hospital (Turin, Italy) were enrolled, including 31 PAPS, 25 SAPS, 27SLE patients without aPL, 29 aPLcarriers (mean age 48.3±13.3 years, 76% female). Nineteen subjects were also recruited as healthy controls (HCs). Gene expression was evaluated by RT-PCR for the following genes: IFI6, IFI44, IFI44L, MX1,IFI27, OAS1 and RSAD2. Normalized gene expression levels (Z-scores) were averaged into a global IFN signature (IFN score). Differences were measured by Kruskal-Wallis tests and associations among genes were studied by cluster and correspondence analyses. Correlations among genes were plotted by network analyses.
Results: An overall activation of ISG was noted across APS subsets, but differences were noted among genes. Whereas some ISG were already upregulated in the aPL positive group compared to HCs (IFI44, IFI44L, MX1, IFI27, OAS1 and RSAD2, all p< 0.050), other ISG were only in increased SLE (IFI6), MX1 differed between SLE and SAPS, and IFI27 and OAS1 showed differences between PAPS and SAPS. The composite IFN score revealed quantitative differences in the IFN pathway activation across APS subsets, being elevated in aPL carriers/PAPS groups compared to HCs (both p< 0.050) and increasing in SAPS (p< 0.010) and SLE (p< 0.001) groups. Network analyses (Figure 1A) revealed qualitative differences in the gene-gene correlation networks: (i) weaker structures were found in HCs and aPL carriers, compared to stronger and higher-degree networks in SAPS and SLE groups; and (ii) the influence of each node was different across groups. Unsupervised cluster analysis identified 3 clusters (I to III) based on ISG patterns (figure 1B). Clusters usage differed among APS subsets, thus correlating clinical status (Figure 1C). Distinct groups of ISG positively correlate to aPS/PT IgG titre in aPL carriers and PAPS groups (all rho >0.500), whereas no associations were retrieved in SAPS or SLE. No associations with thrombotic events were observed, although IFN composite score and several ISG correlate with the number of thrombotic recurrences under anticoagulation (all rho >0.400).
Conclusion: An overall IFN pathway activation has been observed in aPL positive patients and across all APS subsets. Qualitative and quantitative differences across the APS spectrum can be identified, leading tothe identification of distinct IFN signatures with different clinical value.
 Figure 1.
Figure 1.1A) Network analyses of the interferon regulated genes among different subsets; 1B and 1C) results of cluster analysis and correlations with different clinical status.
Disclosures: I. Cecchi, None; M. Radin, None; A. Barinotti, None; S. Foddai, None; E. Rubini, None; A. Suárez, None; S. Baldovino, None; E. Menegatti, None; D. Roccatello, None; S. Sciascia, None; J. Rodríguez-Carrio, None.

