Back
Abstract Session
Systemic lupus erythematosus (SLE)
Session: Abstracts: SLE – Animal Models (1663–1666)
1666: Differentiation of Injury-associated Macrophages in Lupus Kidneys Is Conserved in Humans and Lupus Mouse Models
Monday, November 14, 2022
11:15 AM – 11:25 AM Eastern Time
Location: Room 114 Nutter Theatre
.png)
Paul Hoover, MD, PhD
Brigham and Women's Hospital
Boston, MA, United States
Presenting Author(s)
Paul Hoover1, David Lieb2, Stephen Li2, Chirag Raparia3, Accelerating Medicines Partnership SLE/RA4, Arnon Arazi5, Nir Hacohen2 and Anne Davidson6, 1Brigham and Women's Hospital, Boston, MA, 2Broad Institute, Cambridge, MA, 3Donald and Barbara Zucker School of Medicine At Hofstra/Northwell, Shoreham, NY, 4NIH, Bethesda, MD, 5Feinstein Institutes for Medical Research, Melrose, MA, 6Feinstein Institutes for Medical Research, Manhasset, NY
Background/Purpose: Infiltrating monocytes acquire functions that support kidney remodeling in response to tissue damage in lupus nephritis. This process of monocyte differentiation has been difficult to study due to the inability to characterize immune cells from small human kidney biopsies or to accurately use mouse models to investigate mechanisms relevant to human disease. We previously reported on the identification of comparable kidney macrophage states from 24 patients with lupus nephritis and 4 common lupus mouse models using single cell RNA seq. Here, we compared intrarenal myeloid cells from 155 patients from AMP Phase 2 and our 4 lupus mouse models to discover that injury-associated macrophages and their differentiation from infiltrating monocytes is comparable in humans and mice.
Methods: For humans, we analyzed ~25,000 intrarenal myeloid cell transcriptomes in collaboration with AMP-SLE. For mice, we sorted CD45+ cells from dissociated mouse kidneys with spontaneous adaptive-driven autoimmunity (NZB/W & MRL/lpr) and TLR7-overexpression innate-driven autoimmunity (Sle1.Yaa & NZW/BXSB) in early and nephritic disease. We profiled single cells using 10x Genomics, analyzed ~10,000 transcriptomes with >500 genes and UMIs using Seurat 3.0, and performed Louvain-based clustering. We developed an antibody panel based on the top differentially expressed genes for mouse myeloid clusters for flow-cytometry. To identify similar mouse and human myeloid states we applied classifiers trained on our mouse data to AMP myeloid data. We used partition-based graph abstraction to compare cellular trajectories from analogous human and mouse myeloid clusters.
Results: Injury-associated Clec4n+ and C3ar1+ macrophage populations emerged in nephritic kidneys from all 4 mouse strains. In humans, we identified comparable macrophage populations that correlated with active kidney injury (Fig. 1). These injury-associated states adopted a similar differentiation path in mice and humans that derived from an analogous infiltrating monocyte (Fig. 2). Similar gene programs and transcription factors shifted expression over this differentiation path and culminated in genes for lipid processing and phagocytosis in the injury-associated states.
Conclusion: Our studies of 155 patients and 4 common lupus mouse models identified comparable injury-associated macrophage states in lupus nephritis. These cells were enriched for lipid processing and phagocytosis gene modules and differentiated through a conserved set of cellular states that expressed similar gene programs and transcription factors. These injury-associated states may be critical for remodeling injured lupus kidneys and can now be studied in future mouse work that will accurately reflect this aspect of human lupus nephritis.
.jpg) Fig. 1 Phagocytic macs and a subset of reparative macs correlate with kidney injury in human lupus nephritis. LEFT. ~25,000 intrarenal myeloid cells collected from 155 lupus nephritis patient biopsies. RIGHT. Association of cells with kidney injury (activity index) by covarying neighborhood analysis. Analysis with AMP-SLE consortium.
Fig. 1 Phagocytic macs and a subset of reparative macs correlate with kidney injury in human lupus nephritis. LEFT. ~25,000 intrarenal myeloid cells collected from 155 lupus nephritis patient biopsies. RIGHT. Association of cells with kidney injury (activity index) by covarying neighborhood analysis. Analysis with AMP-SLE consortium.
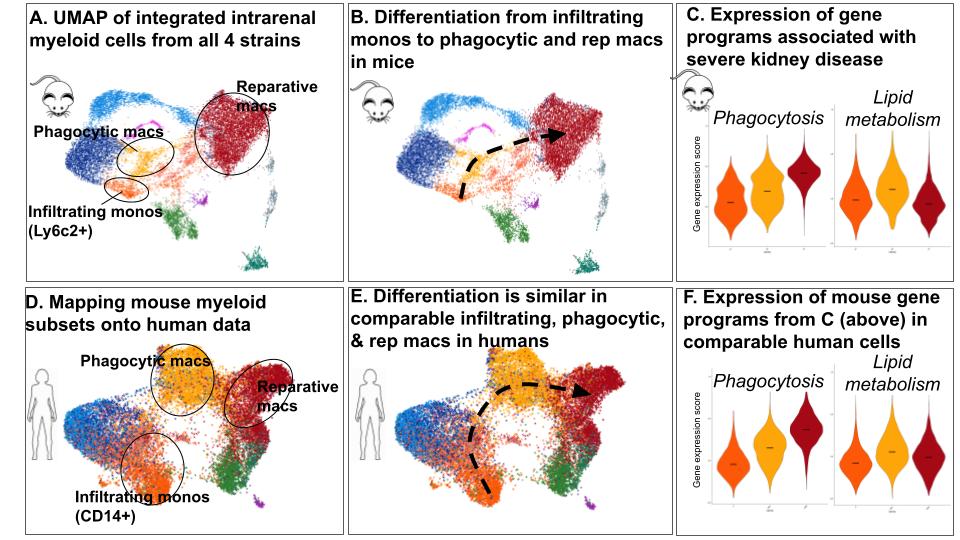 Fig. 2. Injury-associated macrophages differentiated through a conserved set of cellular states. Identification of comparable macrophage states in mouse and human lupus nephritis, A & D. Infiltrating and injury-associated-states adopt the same differentiation path, B & E. Average expression of genes for phagocytosis (41 genes) and lipid metabolism (12 genes) were monitored over this differentiation path, C & F.
Fig. 2. Injury-associated macrophages differentiated through a conserved set of cellular states. Identification of comparable macrophage states in mouse and human lupus nephritis, A & D. Infiltrating and injury-associated-states adopt the same differentiation path, B & E. Average expression of genes for phagocytosis (41 genes) and lipid metabolism (12 genes) were monitored over this differentiation path, C & F.
Disclosures: P. Hoover, Sanofi, Third Rock Ventures; D. Lieb, None; S. Li, None; C. Raparia, None; A. SLE/RA, None; A. Arazi, None; N. Hacohen, DangerBio/Related Sciences; A. Davidson, None.
Background/Purpose: Infiltrating monocytes acquire functions that support kidney remodeling in response to tissue damage in lupus nephritis. This process of monocyte differentiation has been difficult to study due to the inability to characterize immune cells from small human kidney biopsies or to accurately use mouse models to investigate mechanisms relevant to human disease. We previously reported on the identification of comparable kidney macrophage states from 24 patients with lupus nephritis and 4 common lupus mouse models using single cell RNA seq. Here, we compared intrarenal myeloid cells from 155 patients from AMP Phase 2 and our 4 lupus mouse models to discover that injury-associated macrophages and their differentiation from infiltrating monocytes is comparable in humans and mice.
Methods: For humans, we analyzed ~25,000 intrarenal myeloid cell transcriptomes in collaboration with AMP-SLE. For mice, we sorted CD45+ cells from dissociated mouse kidneys with spontaneous adaptive-driven autoimmunity (NZB/W & MRL/lpr) and TLR7-overexpression innate-driven autoimmunity (Sle1.Yaa & NZW/BXSB) in early and nephritic disease. We profiled single cells using 10x Genomics, analyzed ~10,000 transcriptomes with >500 genes and UMIs using Seurat 3.0, and performed Louvain-based clustering. We developed an antibody panel based on the top differentially expressed genes for mouse myeloid clusters for flow-cytometry. To identify similar mouse and human myeloid states we applied classifiers trained on our mouse data to AMP myeloid data. We used partition-based graph abstraction to compare cellular trajectories from analogous human and mouse myeloid clusters.
Results: Injury-associated Clec4n+ and C3ar1+ macrophage populations emerged in nephritic kidneys from all 4 mouse strains. In humans, we identified comparable macrophage populations that correlated with active kidney injury (Fig. 1). These injury-associated states adopted a similar differentiation path in mice and humans that derived from an analogous infiltrating monocyte (Fig. 2). Similar gene programs and transcription factors shifted expression over this differentiation path and culminated in genes for lipid processing and phagocytosis in the injury-associated states.
Conclusion: Our studies of 155 patients and 4 common lupus mouse models identified comparable injury-associated macrophage states in lupus nephritis. These cells were enriched for lipid processing and phagocytosis gene modules and differentiated through a conserved set of cellular states that expressed similar gene programs and transcription factors. These injury-associated states may be critical for remodeling injured lupus kidneys and can now be studied in future mouse work that will accurately reflect this aspect of human lupus nephritis.
.jpg) Fig. 1 Phagocytic macs and a subset of reparative macs correlate with kidney injury in human lupus nephritis. LEFT. ~25,000 intrarenal myeloid cells collected from 155 lupus nephritis patient biopsies. RIGHT. Association of cells with kidney injury (activity index) by covarying neighborhood analysis. Analysis with AMP-SLE consortium.
Fig. 1 Phagocytic macs and a subset of reparative macs correlate with kidney injury in human lupus nephritis. LEFT. ~25,000 intrarenal myeloid cells collected from 155 lupus nephritis patient biopsies. RIGHT. Association of cells with kidney injury (activity index) by covarying neighborhood analysis. Analysis with AMP-SLE consortium. Fig. 2. Injury-associated macrophages differentiated through a conserved set of cellular states. Identification of comparable macrophage states in mouse and human lupus nephritis, A & D. Infiltrating and injury-associated-states adopt the same differentiation path, B & E. Average expression of genes for phagocytosis (41 genes) and lipid metabolism (12 genes) were monitored over this differentiation path, C & F.
Fig. 2. Injury-associated macrophages differentiated through a conserved set of cellular states. Identification of comparable macrophage states in mouse and human lupus nephritis, A & D. Infiltrating and injury-associated-states adopt the same differentiation path, B & E. Average expression of genes for phagocytosis (41 genes) and lipid metabolism (12 genes) were monitored over this differentiation path, C & F.Disclosures: P. Hoover, Sanofi, Third Rock Ventures; D. Lieb, None; S. Li, None; C. Raparia, None; A. SLE/RA, None; A. Arazi, None; N. Hacohen, DangerBio/Related Sciences; A. Davidson, None.

