Back
Poster Session E - Tuesday Afternoon
E0306 - Oversampling in Machine Learning for Patient Risk Stratification for Acute Lower Gastrointestinal Bleeding
Tuesday, October 25, 2022
3:00 PM – 5:00 PM ET
Location: Crown Ballroom

Jonathan J. Ho, MD
Warren Alpert Medical School of Brown University
Providence, RI
Presenting Author(s)
Jonathan Ho, MD1, George M. Hanna, MD2, Amber Charoen, MD3, Fadlallah Habr, MD4
1Warren Alpert Medical School of Brown University , Providence, RI; 2Warren Alpert Medical School of Brown University, Providence, RI; 3Brown University, Cranston, RI; 4Warren Alpert Medical School of Brown University, East Providence, RI
Introduction: Lower gastrointestinal bleeding (LGIB) is a common cause of hospital admissions and can lead to hospital-based interventions that consume a significant amount of medical resources. However, only a minority of cases are high-risk and result in significant morbidity and mortality. We present an oversampling method to help with rebalancing for machine learning modeling for triaging in LGIB when there is significant imbalance between high risk (HR) and low risk (LR) patients.
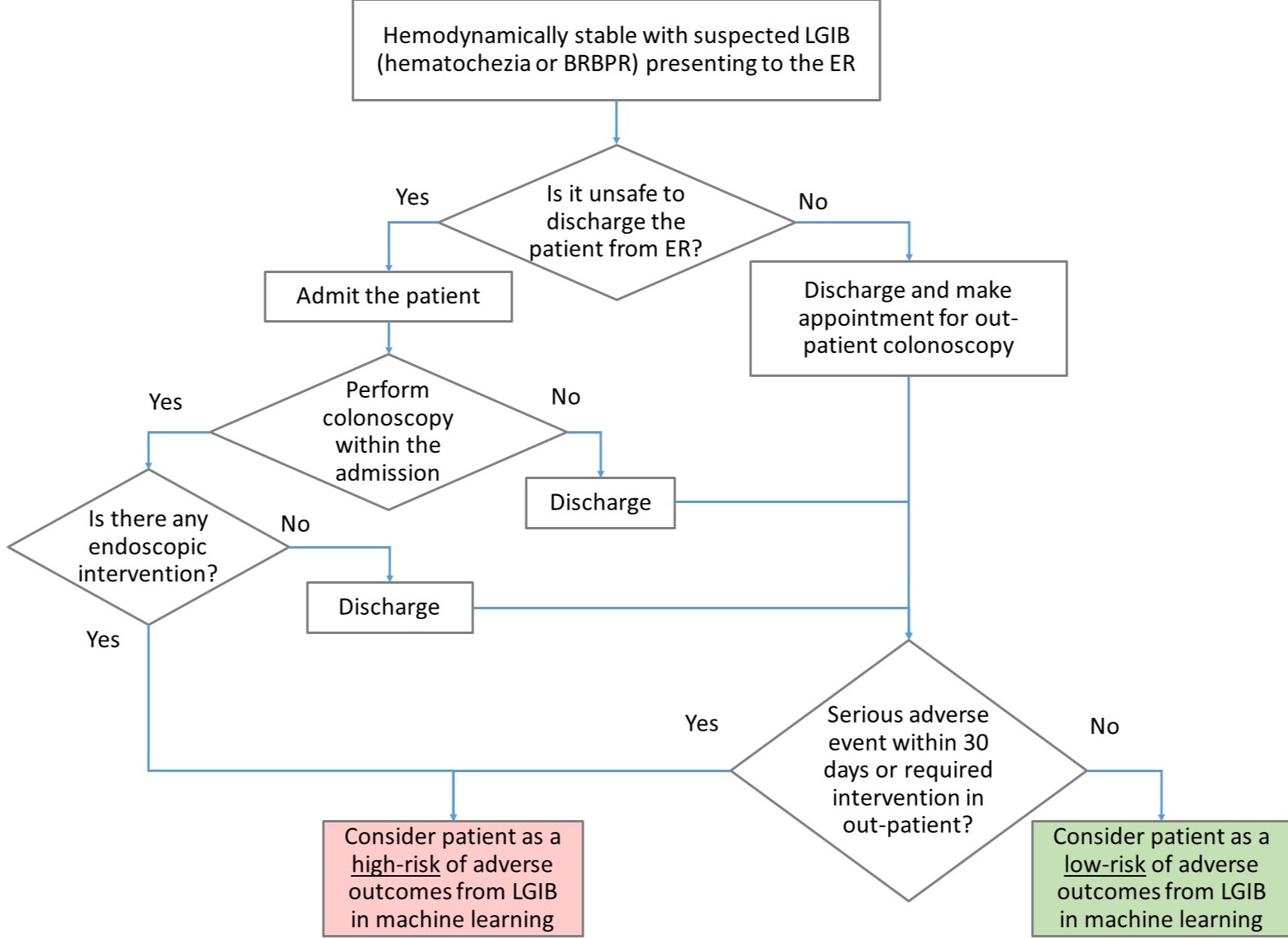
Methods: From retrospective data, hemodynamically stable patients with suspected LGIB were labeled into HR or LR groups (Figure 1). Risk factors associated with LGIB (e.g. age, sex, blood pressure, hemoglobin) were included as predictors. The dataset was divided into 80% for training and 20% for testing. Two machine learning models (stepwise logistic regression and decision trees) were applied to the training data to create predictive models. Then, the training and testing performances were evaluated using standard performance metrics (e.g. sensitivity, specificity, and F1).
Results: 1414 records were reviewed. On average, patients were 61 years old and 48.8% were male. The average systolic blood pressure was 138 mmHg and diastolic was 78.0 mmHg with an average pulse of 82.0. The average laboratory values were 13.2 g/dL for hemoglobin (Hb), 16.0 mg/dL for BUN, 0.83 mg/dL for creatinine, 1.1 for INR, and 227.0 × 10e9/L for platelets. Among these patients, 14% were on anticoagulants, 4.3% were on antiplatelet agents, and 14.9% took NSAIDs. There were 69 HR patients and 1345 LR patients. Among the included factors, age, blood pressure, pulse, BUN, Hb, INR, quartile of transfusions, and being on antiplatelet agents were statistically different between the 2 risk groups. During training the decision tree model showed excellent specificity (0.985) and negative predictive value (NPV) (0.9088) among 586 cases. In the testing phase, specificity was 0.982 and NPV was 0.960 among 12 cases. Sensitivity dropped from 0.908 in the training phase to 0.083 in the testing phase; similarly, the F1 dropped from 0.945 to 0.111. The logistic regression model had a sensitivity of 0.691 that dropped to 0.583 and specificity from 0.723 to 0.752; the F1 dropped from 0.709 to 0.163.
Discussion: Logistic regression did not perform as well as decision trees in training; however, it can generalize better to unseen data. A larger dataset with more HR cases would potentially reduce the overfitting issue and provide a more accurate predictive model.
Disclosures:
Jonathan Ho, MD1, George M. Hanna, MD2, Amber Charoen, MD3, Fadlallah Habr, MD4. E0306 - Oversampling in Machine Learning for Patient Risk Stratification for Acute Lower Gastrointestinal Bleeding, ACG 2022 Annual Scientific Meeting Abstracts. Charlotte, NC: American College of Gastroenterology.
1Warren Alpert Medical School of Brown University , Providence, RI; 2Warren Alpert Medical School of Brown University, Providence, RI; 3Brown University, Cranston, RI; 4Warren Alpert Medical School of Brown University, East Providence, RI
Introduction: Lower gastrointestinal bleeding (LGIB) is a common cause of hospital admissions and can lead to hospital-based interventions that consume a significant amount of medical resources. However, only a minority of cases are high-risk and result in significant morbidity and mortality. We present an oversampling method to help with rebalancing for machine learning modeling for triaging in LGIB when there is significant imbalance between high risk (HR) and low risk (LR) patients.
Methods: From retrospective data, hemodynamically stable patients with suspected LGIB were labeled into HR or LR groups (Figure 1). Risk factors associated with LGIB (e.g. age, sex, blood pressure, hemoglobin) were included as predictors. The dataset was divided into 80% for training and 20% for testing. Two machine learning models (stepwise logistic regression and decision trees) were applied to the training data to create predictive models. Then, the training and testing performances were evaluated using standard performance metrics (e.g. sensitivity, specificity, and F1).
Results: 1414 records were reviewed. On average, patients were 61 years old and 48.8% were male. The average systolic blood pressure was 138 mmHg and diastolic was 78.0 mmHg with an average pulse of 82.0. The average laboratory values were 13.2 g/dL for hemoglobin (Hb), 16.0 mg/dL for BUN, 0.83 mg/dL for creatinine, 1.1 for INR, and 227.0 × 10e9/L for platelets. Among these patients, 14% were on anticoagulants, 4.3% were on antiplatelet agents, and 14.9% took NSAIDs. There were 69 HR patients and 1345 LR patients. Among the included factors, age, blood pressure, pulse, BUN, Hb, INR, quartile of transfusions, and being on antiplatelet agents were statistically different between the 2 risk groups. During training the decision tree model showed excellent specificity (0.985) and negative predictive value (NPV) (0.9088) among 586 cases. In the testing phase, specificity was 0.982 and NPV was 0.960 among 12 cases. Sensitivity dropped from 0.908 in the training phase to 0.083 in the testing phase; similarly, the F1 dropped from 0.945 to 0.111. The logistic regression model had a sensitivity of 0.691 that dropped to 0.583 and specificity from 0.723 to 0.752; the F1 dropped from 0.709 to 0.163.
Discussion: Logistic regression did not perform as well as decision trees in training; however, it can generalize better to unseen data. A larger dataset with more HR cases would potentially reduce the overfitting issue and provide a more accurate predictive model.

Figure: Figure 1. Algorithm for classifying patients with low-risk or high-risk LGIB
BRBPR = Bright red blood per rectum, ER = Emergency room, LGIB = Lower gastrointestinal bleeding
BRBPR = Bright red blood per rectum, ER = Emergency room, LGIB = Lower gastrointestinal bleeding
Disclosures:
Jonathan Ho indicated no relevant financial relationships.
George Hanna indicated no relevant financial relationships.
Amber Charoen indicated no relevant financial relationships.
Fadlallah Habr indicated no relevant financial relationships.
Jonathan Ho, MD1, George M. Hanna, MD2, Amber Charoen, MD3, Fadlallah Habr, MD4. E0306 - Oversampling in Machine Learning for Patient Risk Stratification for Acute Lower Gastrointestinal Bleeding, ACG 2022 Annual Scientific Meeting Abstracts. Charlotte, NC: American College of Gastroenterology.
