Back

Poster Session E - Tuesday Afternoon
Category: Colon
E0105 - Multimodal Analysis of cfDNA Methylation Sequencing Improves Early Colon Cancer Detection
Tuesday, October 25, 2022
3:00 PM – 5:00 PM ET
Location: Crown Ballroom

Has Audio

Shivani Mahajan, PhD
Helio Genomics
Irvine, CA
Presenting Author(s)
Shivani Mahajan, PhD, Itai Pinkoviezky, PhD, Jianfeng Xu, PhD, Maxime Gallant, PhD, Allison Sorg, , David Taggart, PhD
Helio Genomics, Irvine, CA
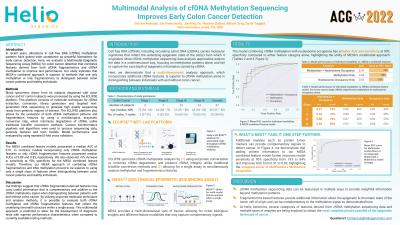
Introduction: In recent years, alterations in cell free DNA (cfDNA) methylation patterns have gained wide acceptance as powerful biomarkers for early cancer detection. Here, we evaluate a Multimodal Epigenetic Sequencing Assay (MESA) for colon cancer detection that combines features derived from both cfDNA fragmentomics and cfDNA methylation to improve test performance. Our study indicates that MESA’s combined approach fragmentomics to distinguish between colon cancer patients and healthy individuals.
Methods: Blood specimens drawn from 64 subjects diagnosed with colon cancer and 67 control subjects were processed by using the ECLIPSE platform. This platform consists of molecular techniques for cfDNA extraction, conversion, library generation and targeted next-generation DNA sequencing to generate high quality sequencing reads from genomic regions of interest. The ECLIPSE platform also allows for the evaluation of both cfDNA methylation patterns and fragmentation features by using a non-disruptive, enzymatic conversion step which minimizes degradation of cfDNA, unlike traditional bisulfite conversion methods. Custom bioinformatics pipelines and algorithms were used to process sequencing data, generate features and train models. Model performance was evaluated by using repeated 5-fold cross validation.
Results: The MESA combined feature models possessed a median AUC of 0.91. In contrast, models incorporating only cfDNA methylation features or only cfDNA fragmentation features possessed median AUCs of 0.89 and 0.83, respectively. We also observed > 5% increase in sensitivity at 90% specificity for the MESA combined feature models. Therefore, our MESA approach of combining cfDNA fragmentomics and DNA methylation proved to be superior to using only a single class of features when distinguishing between colon cancer patients and healthy individuals.
Discussion: Our findings suggest that cfDNA fragmentation-derived features may carry useful information that is complementary and additive to the cfDNA methylation signal when distinguishing between patients with and without colon cancer. By utilizing improved molecular techniques and analysis methods, it is possible to evaluate both cfDNA methylation and cfDNA fragmentation features that reflect the underlying chromatin structure within a single assay. This multimodal approach is predicted to allow for the development of diagnostic tests with superior performance characteristics when compared to currently available testing methods.
Disclosures:
Shivani Mahajan, PhD, Itai Pinkoviezky, PhD, Jianfeng Xu, PhD, Maxime Gallant, PhD, Allison Sorg, , David Taggart, PhD. E0105 - Multimodal Analysis of cfDNA Methylation Sequencing Improves Early Colon Cancer Detection, ACG 2022 Annual Scientific Meeting Abstracts. Charlotte, NC: American College of Gastroenterology.
Helio Genomics, Irvine, CA
Introduction: In recent years, alterations in cell free DNA (cfDNA) methylation patterns have gained wide acceptance as powerful biomarkers for early cancer detection. Here, we evaluate a Multimodal Epigenetic Sequencing Assay (MESA) for colon cancer detection that combines features derived from both cfDNA fragmentomics and cfDNA methylation to improve test performance. Our study indicates that MESA’s combined approach fragmentomics to distinguish between colon cancer patients and healthy individuals.
Methods: Blood specimens drawn from 64 subjects diagnosed with colon cancer and 67 control subjects were processed by using the ECLIPSE platform. This platform consists of molecular techniques for cfDNA extraction, conversion, library generation and targeted next-generation DNA sequencing to generate high quality sequencing reads from genomic regions of interest. The ECLIPSE platform also allows for the evaluation of both cfDNA methylation patterns and fragmentation features by using a non-disruptive, enzymatic conversion step which minimizes degradation of cfDNA, unlike traditional bisulfite conversion methods. Custom bioinformatics pipelines and algorithms were used to process sequencing data, generate features and train models. Model performance was evaluated by using repeated 5-fold cross validation.
Results: The MESA combined feature models possessed a median AUC of 0.91. In contrast, models incorporating only cfDNA methylation features or only cfDNA fragmentation features possessed median AUCs of 0.89 and 0.83, respectively. We also observed > 5% increase in sensitivity at 90% specificity for the MESA combined feature models. Therefore, our MESA approach of combining cfDNA fragmentomics and DNA methylation proved to be superior to using only a single class of features when distinguishing between colon cancer patients and healthy individuals.
Discussion: Our findings suggest that cfDNA fragmentation-derived features may carry useful information that is complementary and additive to the cfDNA methylation signal when distinguishing between patients with and without colon cancer. By utilizing improved molecular techniques and analysis methods, it is possible to evaluate both cfDNA methylation and cfDNA fragmentation features that reflect the underlying chromatin structure within a single assay. This multimodal approach is predicted to allow for the development of diagnostic tests with superior performance characteristics when compared to currently available testing methods.
Disclosures:
Shivani Mahajan: Freenome – Intellectual Property/Patents, previously employed at Freenome, Stock Options. Helio Genomics – Employee.
Itai Pinkoviezky: Craneware – Employee. Helio Genomics – Employee.
Jianfeng Xu: Helio Genomics – Employee.
Maxime Gallant: Helio Genomics – Employee, Intellectual Property/Patents, Stock Options.
Allison Sorg: Helio Genomics – Employee.
David Taggart: Helio Genomics – Employee.
Shivani Mahajan, PhD, Itai Pinkoviezky, PhD, Jianfeng Xu, PhD, Maxime Gallant, PhD, Allison Sorg, , David Taggart, PhD. E0105 - Multimodal Analysis of cfDNA Methylation Sequencing Improves Early Colon Cancer Detection, ACG 2022 Annual Scientific Meeting Abstracts. Charlotte, NC: American College of Gastroenterology.
